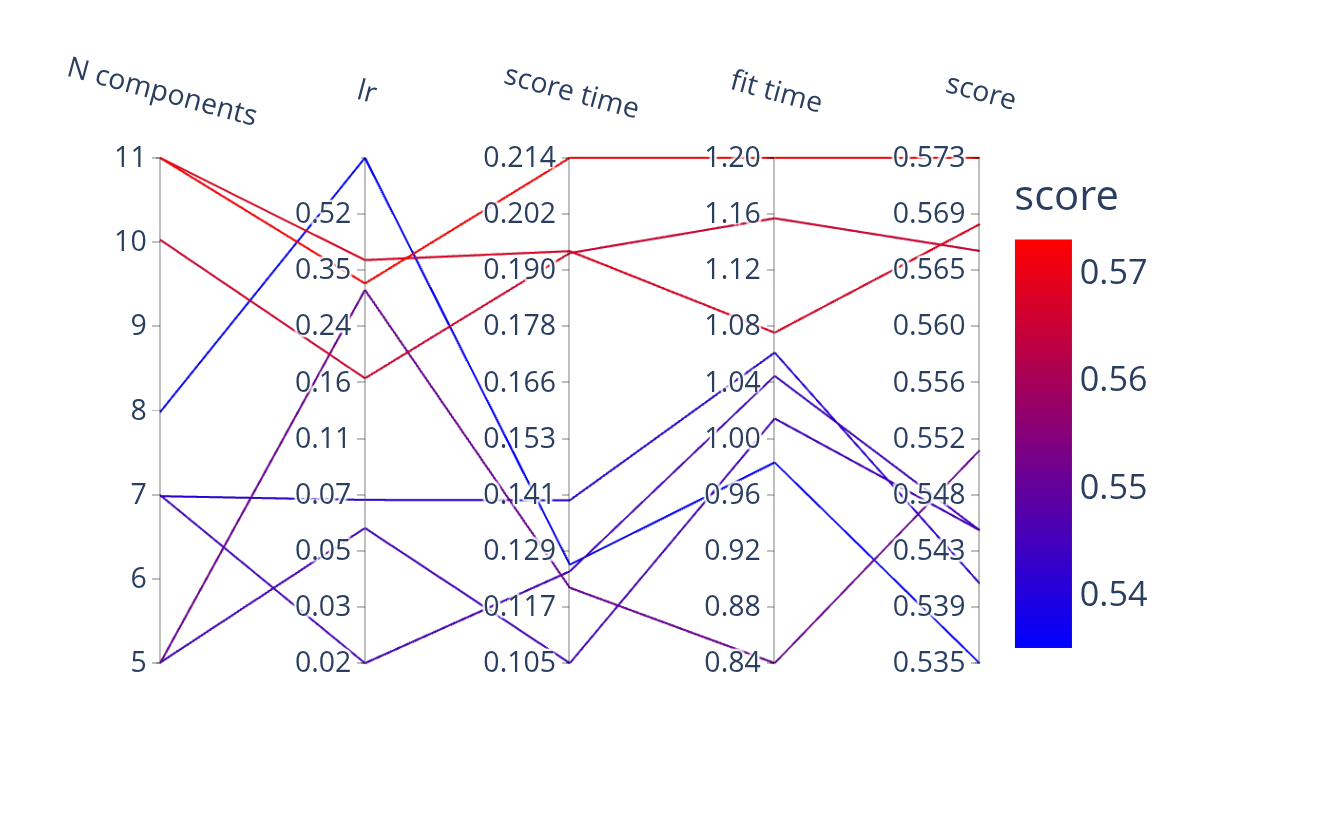
pipe = Pipeline([("dim_reduction", PCA()), ("regressor", Ridge())])
grid = [
{
"dim_reduction": [PCA()],
"dim_reduction__n_components": [10, 20, 30],
"regressor": [Ridge()],
"regressor__alpha": loguniform(0.1, 10.0),
},
{
"dim_reduction": [SelectKBest()],
"dim_reduction__k": [10, 20, 30],
"regressor": [Ridge()],
"regressor__alpha": loguniform(0.1, 10.0),
},
{
"dim_reduction": [PCA()],
"dim_reduction__n_components": [10, 20, 30],
"regressor": [RandomForestClassifier()],
"regressor__n_estimators": loguniform(20, 200),
},
{
"dim_reduction": [SelectKBest()],
"dim_reduction__k": [10, 20, 30],
"regressor": [RandomForestClassifier()],
"regressor__n_estimators": loguniform(20, 200),
},
]
model = RandomizedSearchCV(pipe, grid)