EDF Talk: Skrub
Machine learning with dataframes
Inria P16
2025-09-16
Boost your productivity with skrub!
Skrub simplifies many tedious data preparation operations
skrub compatibility
- skrub is fully compatible with pandas and polars
- skrub transformers are fully compatible with scikit-learn
An example pipeline
- Gather some data
- Explore the data
- Preprocess the data
- Perform feature engineering
- Build a scikit-learn pipeline
- ???
- Profit?
Exploring the data
import pandas as pd
import matplotlib.pyplot as plt
import skrub
dataset = skrub.datasets.fetch_employee_salaries()
employees, salaries = dataset.X, dataset.y
df = pd.DataFrame(employees)
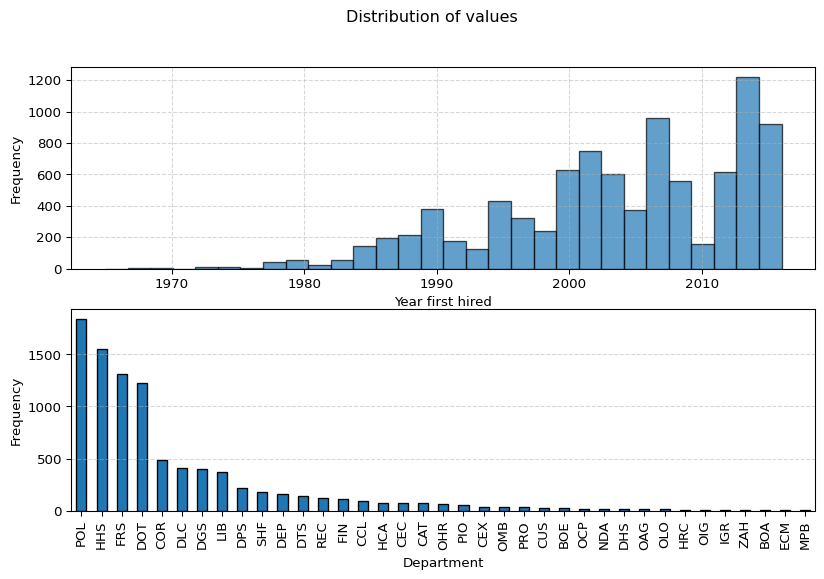
# Plot the distribution of the numerical values using a histogram
fig, axs = plt.subplots(2,1, figsize=(10, 6))
ax1, ax2 = axs
ax1.hist(df['year_first_hired'], bins=30, edgecolor='black', alpha=0.7)
ax1.set_xlabel('Year first hired')
ax1.set_ylabel('Frequency')
ax1.grid(True, linestyle='--', alpha=0.5)
# Count the frequency of each category
category_counts = df['department'].value_counts()
# Create a bar plot
category_counts.plot(kind='bar', edgecolor='black', ax=ax2)
# Add labels and title
ax2.set_xlabel('Department')
ax2.set_ylabel('Frequency')
ax2.grid(True, linestyle='--', axis='y', alpha=0.5) # Add grid lines for y-axis
fig.suptitle("Distribution of values")
# Show the plot
plt.show()Exploring the data
Exploring the data with skrub
Main features:
- Obtain high-level statistics about the data
- Explore the distribution of values and find outliers
- Discover highly correlated columns
- Export and share the report as an
htmlfile
Data cleaning with pandas/polars: setup
import pandas as pd
import numpy as np
data = {
'Constant int': [1, 1, 1], # Single unique value
'B': [2, 3, 2], # Multiple unique values
'Constant str': ['x', 'x', 'x'], # Single unique value
'D': [4, 5, 6], # Multiple unique values
'All nan': [np.nan, np.nan, np.nan], # All missing values
'All empty': ['', '', ''], # All empty strings
'Date': ['01/01/2023', '02/01/2023', '03/01/2023'],
}
df_pd = pd.DataFrame(data)
display(df_pd)| Constant int | B | Constant str | D | All nan | All empty | Date | |
|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | x | 4 | NaN | 01/01/2023 | |
| 1 | 1 | 3 | x | 5 | NaN | 02/01/2023 | |
| 2 | 1 | 2 | x | 6 | NaN | 03/01/2023 |
import polars as pl
import numpy as np
data = {
'Constant int': [1, 1, 1], # Single unique value
'B': [2, 3, 2], # Multiple unique values
'Constant str': ['x', 'x', 'x'], # Single unique value
'D': [4, 5, 6], # Multiple unique values
'All nan': [np.nan, np.nan, np.nan], # All missing values
'All empty': ['', '', ''], # All empty strings
'Date': ['01/01/2023', '02/01/2023', '03/01/2023'],
}
df_pl = pl.DataFrame(data)
display(df_pl)| Constant int | B | Constant str | D | All nan | All empty | Date |
|---|---|---|---|---|---|---|
| i64 | i64 | str | i64 | f64 | str | str |
| 1 | 2 | "x" | 4 | NaN | "" | "01/01/2023" |
| 1 | 3 | "x" | 5 | NaN | "" | "02/01/2023" |
| 1 | 2 | "x" | 6 | NaN | "" | "03/01/2023" |
Nulls, datetimes, constant columns with pandas/polars
# Parse the datetime strings with a specific format
df_pd['Date'] = pd.to_datetime(df_pd['Date'], format='%d/%m/%Y')
# Drop columns with only a single unique value
df_pd_cleaned = df_pd.loc[:, df_pd.nunique(dropna=True) > 1]
# Function to drop columns with only missing values or empty strings
def drop_empty_columns(df):
# Drop columns with only missing values
df_cleaned = df.dropna(axis=1, how='all')
# Drop columns with only empty strings
empty_string_cols = df_cleaned.columns[df_cleaned.eq('').all()]
df_cleaned = df_cleaned.drop(columns=empty_string_cols)
return df_cleaned
# Apply the function to the DataFrame
df_pd_cleaned = drop_empty_columns(df_pd_cleaned)# Parse the datetime strings with a specific format
df_pl = df_pl.with_columns([
pl.col("Date").str.strptime(pl.Date, "%d/%m/%Y", strict=False).alias("Date")
])
# Drop columns with only a single unique value
df_pl_cleaned = df_pl.select([
col for col in df_pl.columns if df_pl[col].n_unique() > 1
])
# Import selectors for dtype selection
import polars.selectors as cs
# Drop columns with only missing values or only empty strings
def drop_empty_columns(df):
all_nan = df.select(
[
col for col in df.select(cs.numeric()).columns if
df [col].is_nan().all()
]
).columns
all_empty = df.select(
[
col for col in df.select(cs.string()).columns if
(df[col].str.strip_chars().str.len_chars()==0).all()
]
).columns
to_drop = all_nan + all_empty
return df.drop(to_drop)
df_pl_cleaned = drop_empty_columns(df_pl_cleaned)Data cleaning with skrub.Cleaner
Comparison
Encoding datetime features with pandas/polars
import pandas as pd
data = {
'date': ['2023-01-01 12:34:56', '2023-02-15 08:45:23', '2023-03-20 18:12:45'],
'value': [10, 20, 30]
}
df_pd = pd.DataFrame(data)
datetime_column = "date"
df_pd[datetime_column] = pd.to_datetime(df_pd[datetime_column], errors='coerce')
df_pd['year'] = df_pd[datetime_column].dt.year
df_pd['month'] = df_pd[datetime_column].dt.month
df_pd['day'] = df_pd[datetime_column].dt.day
df_pd['hour'] = df_pd[datetime_column].dt.hour
df_pd['minute'] = df_pd[datetime_column].dt.minute
df_pd['second'] = df_pd[datetime_column].dt.secondimport polars as pl
data = {
'date': ['2023-01-01 12:34:56', '2023-02-15 08:45:23', '2023-03-20 18:12:45'],
'value': [10, 20, 30]
}
df_pl = pl.DataFrame(data)
df_pl = df_pl.with_columns(date=pl.col("date").str.to_datetime())
df_pl = df_pl.with_columns(
year=pl.col("date").dt.year(),
month=pl.col("date").dt.month(),
day=pl.col("date").dt.day(),
hour=pl.col("date").dt.hour(),
minute=pl.col("date").dt.minute(),
second=pl.col("date").dt.second(),
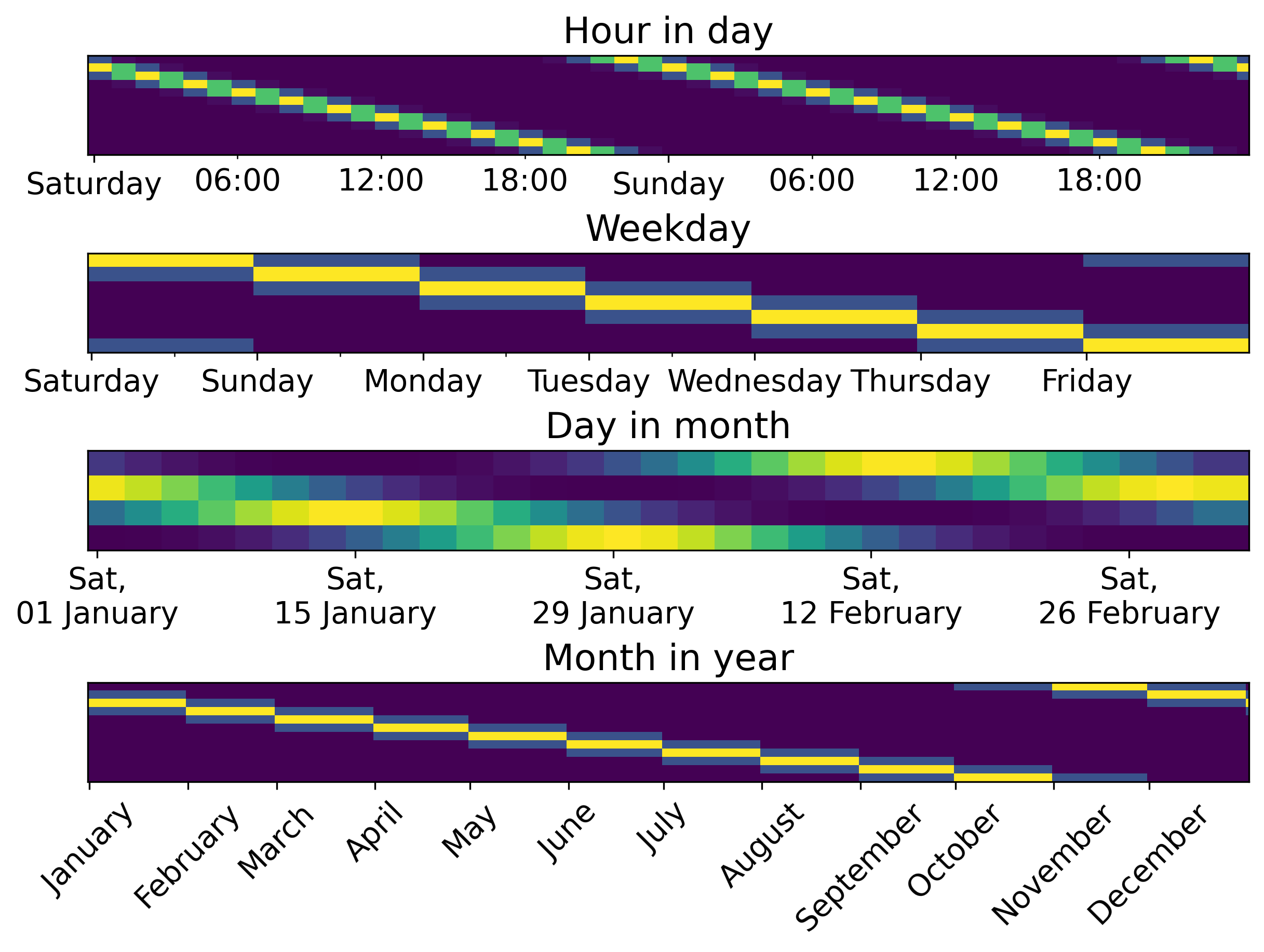
)Adding periodic features with pandas/polars
Encoding datetime features skrub.DatetimeEncoder
shape: (3, 8)
┌───────────┬────────────┬────────────┬────────────┬───────────┬───────────┬───────────┬───────────┐
│ date_year ┆ date_total ┆ date_month ┆ date_month ┆ date_day_ ┆ date_day_ ┆ date_hour ┆ date_hour │
│ --- ┆ _seconds ┆ _circular_ ┆ _circular_ ┆ circular_ ┆ circular_ ┆ _circular ┆ _circular │
│ f32 ┆ --- ┆ 0 ┆ 1 ┆ 0 ┆ 1 ┆ _0 ┆ _1 │
│ ┆ f32 ┆ --- ┆ --- ┆ --- ┆ --- ┆ --- ┆ --- │
│ ┆ ┆ f64 ┆ f64 ┆ f64 ┆ f64 ┆ f64 ┆ f64 │
╞═══════════╪════════════╪════════════╪════════════╪═══════════╪═══════════╪═══════════╪═══════════╡
│ 2023.0 ┆ 1.6726e9 ┆ 0.5 ┆ 0.866025 ┆ 0.207912 ┆ 0.978148 ┆ 1.2246e-1 ┆ -1.0 │
│ ┆ ┆ ┆ ┆ ┆ ┆ 6 ┆ │
│ 2023.0 ┆ 1.6765e9 ┆ 0.866025 ┆ 0.5 ┆ 1.2246e-1 ┆ -1.0 ┆ 0.866025 ┆ -0.5 │
│ ┆ ┆ ┆ ┆ 6 ┆ ┆ ┆ │
│ 2023.0 ┆ 1.6793e9 ┆ 1.0 ┆ 6.1232e-17 ┆ -0.866025 ┆ -0.5 ┆ -1.0 ┆ -1.8370e- │
│ ┆ ┆ ┆ ┆ ┆ ┆ ┆ 16 │
└───────────┴────────────┴────────────┴────────────┴───────────┴───────────┴───────────┴───────────┘What periodic features look like
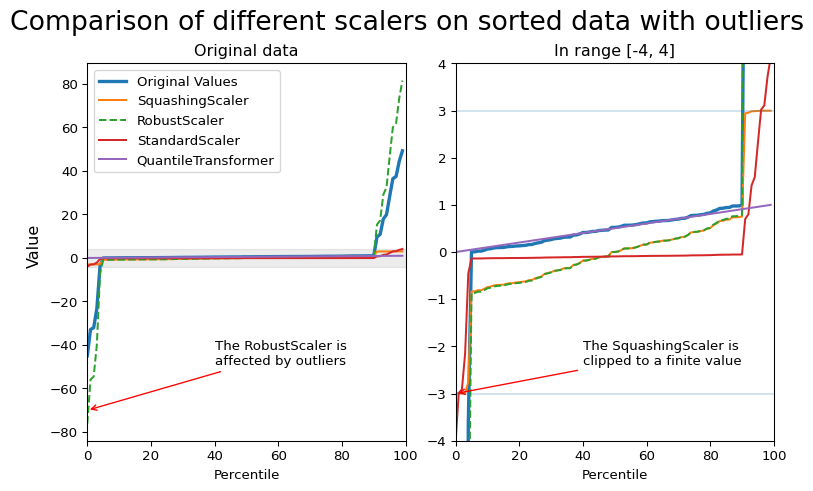
Encoding numerical features with skrub.SquashingScaler
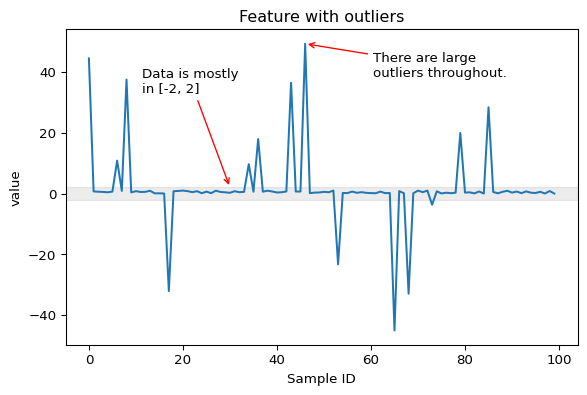
Encoding numerical features with skrub.SquashingScaler
Encoding categorical (string/text) features
Categorical features have a “cardinality”: the number of unique values
- Low cardinality features:
OneHotEncoder - High cardinality features (>40 unique values):
skrub.StringEncoder - Textual features:
skrub.TextEncoderand pretrained models from HuggingFace Hub
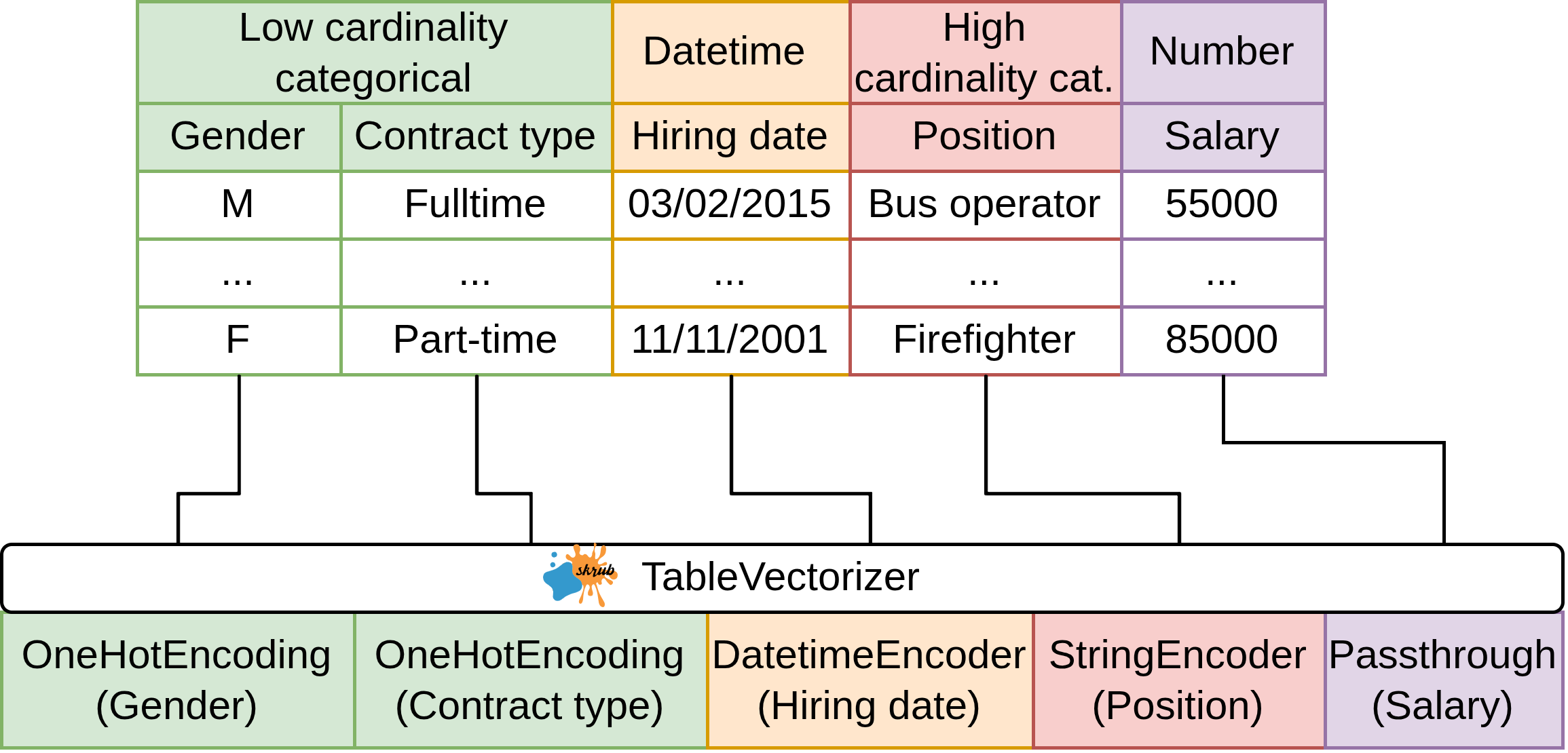
Encoding all the features: TableVectorizer
Encoding all the features: TableVectorizer
Fine-grained column transformations with ApplyToCols
import pandas as pd
from sklearn.compose import make_column_selector as selector
from sklearn.compose import make_column_transformer
from sklearn.preprocessing import StandardScaler, OneHotEncoder
df = pd.DataFrame({"text": ["foo", "bar", "baz"], "number": [1, 2, 3]})
categorical_columns = selector(dtype_include=object)(df)
numerical_columns = selector(dtype_exclude=object)(df)
ct = make_column_transformer(
(StandardScaler(),
numerical_columns),
(OneHotEncoder(handle_unknown="ignore"),
categorical_columns))
transformed = ct.fit_transform(df)
transformedarray([[-1.22474487, 0. , 0. , 1. ],
[ 0. , 1. , 0. , 0. ],
[ 1.22474487, 0. , 1. , 0. ]])Fine-grained column transformations with ApplyToCols
import skrub.selectors as s
from sklearn.pipeline import make_pipeline
from skrub import ApplyToCols
numeric = ApplyToCols(StandardScaler(), cols=s.numeric())
string = ApplyToCols(OneHotEncoder(sparse_output=False), cols=s.string())
transformed = make_pipeline(numeric, string).fit_transform(df)
transformed| text_bar | text_baz | text_foo | number | |
|---|---|---|---|---|
| 0 | 0.0 | 0.0 | 1.0 | -1.224745 |
| 1 | 1.0 | 0.0 | 0.0 | 0.000000 |
| 2 | 0.0 | 1.0 | 0.0 | 1.224745 |
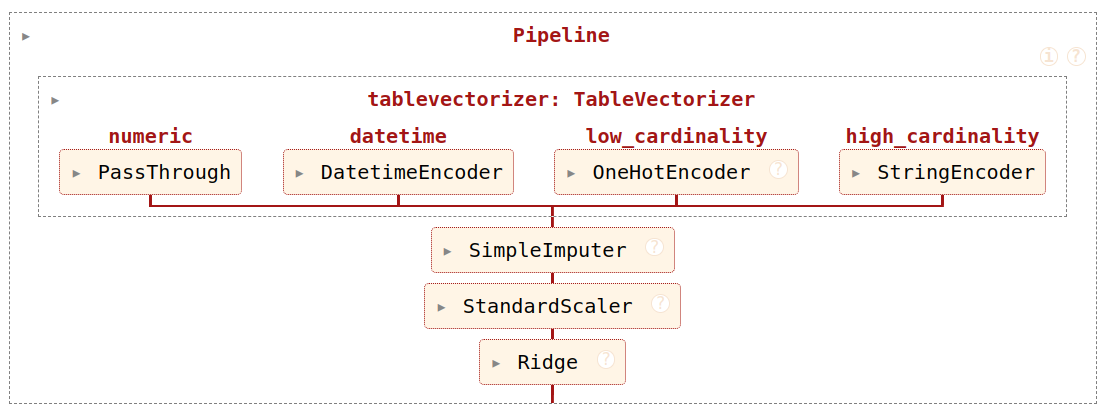
Build a predictive pipeline
Build a predictive pipeline
Build a predictive pipeline
from sklearn.linear_model import Ridge
from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import StandardScaler, OneHotEncoder
from sklearn.impute import SimpleImputer
from sklearn.compose import make_column_selector as selector
from sklearn.compose import make_column_transformer
categorical_columns = selector(dtype_include=object)(employees)
numerical_columns = selector(dtype_exclude=object)(employees)
ct = make_column_transformer(
(StandardScaler(),
numerical_columns),
(OneHotEncoder(handle_unknown="ignore"),
categorical_columns))
model = make_pipeline(ct, SimpleImputer(), Ridge())Build a predictive pipeline with tabular_pipeline

We now have a pipeline!
- Gather some data
- Explore the data
skrub.TableReport
- Pre-process the data
Cleaner,ToDatetime…
- Perform feature engineering
skrub.TableVectorizer,SquashingScaler,TextEncoder,StringEncoder…
- Build a scikit-learn pipeline
tabular_pipeline,sklearn.pipeline.make_pipeline…
- ???
- Profit 📈
What if this is not enough??
What if…
- Your data is spread over multiple tables?
- You want to avoid data leakage?
- You want to tune more than just the hyperparameters of your model?
- You want to guarantee that your pipeline is replayed exactly on new data?
When a normal pipe is not enough…
… the skrub DataOps come to the rescue 🚒
DataOps…
- Extend the
scikit-learnmachinery to complex multi-table operations, and take care of data leakage - Track all operations with a computational graph (a Data Ops plan)
- Allow tuning any operation in the data plan
- Can be persisted and shared easily
How do DataOps work, though?
DataOps wrap around user operations, where user operations are:
- any dataframe operation (e.g., merge, group by, aggregate etc.)
- scikit-learn estimators (a Random Forest, RidgeCV etc.)
- custom user code (load data from a path, fetch from an URL etc.)
Important
DataOps record user operations, so that they can later be replayed in the same order and with the same arguments on unseen data.
DataOps, Plans, learners: oh my!
A
DataOp(singular) wraps a single operation, and can be combined and concatenated with otherDataOps.The Data Ops Plan is a collective name for the directed computational graph that tracks a sequence and combination of
DataOps.The plan can be exported as a standalone object called
learner. Thelearnerworks like a scikit-learn estimator that takes a dictionary of values rather than justXandy.
Starting with the DataOps
X,y,productsrepresent inputs to the pipeline.skrubsplitsXandywhen training.
Building a full data plan
Building a full data plan
Building a full data plan
Inspecting the data plan
Each node:
- Shows a preview of the data resulting from the operation
- Reports the location in the code where the code is defined
- Shows the run time of the node (in the next release)
Exporting the plan in a learner
The data plan can be exported as a learner:
Then, the learner can be pickled …
Hyperparameter tuning in a Data Plan
skrub implements four choose_* functions:
choose_from: select from the given list of optionschoose_int: select an integer within a rangechoose_float: select a float within a rangechoose_bool: select a booloptional: chooses whether to execute the given operation
Tuning in scikit-learn can be complex
pipe = Pipeline([("dim_reduction", PCA()), ("regressor", Ridge())])
grid = [
{
"dim_reduction": [PCA()],
"dim_reduction__n_components": [10, 20, 30],
"regressor": [Ridge()],
"regressor__alpha": loguniform(0.1, 10.0),
},
{
"dim_reduction": [SelectKBest()],
"dim_reduction__k": [10, 20, 30],
"regressor": [Ridge()],
"regressor__alpha": loguniform(0.1, 10.0),
},
{
"dim_reduction": [PCA()],
"dim_reduction__n_components": [10, 20, 30],
"regressor": [RandomForestClassifier()],
"regressor__n_estimators": loguniform(20, 200),
},
{
"dim_reduction": [SelectKBest()],
"dim_reduction__k": [10, 20, 30],
"regressor": [RandomForestClassifier()],
"regressor__n_estimators": loguniform(20, 200),
},
]
model = RandomizedSearchCV(pipe, grid)Tuning with DataOps is simple!
dim_reduction = X.skb.apply(
skrub.choose_from(
{
"PCA": PCA(n_components=skrub.choose_int(10, 30)),
"SelectKBest": SelectKBest(k=skrub.choose_int(10, 30))
}, name="dim_reduction"
)
)
regressor = dim_reduction.skb.apply(
skrub.choose_from(
{
"Ridge": Ridge(alpha=skrub.choose_float(0.1, 10.0, log=True)),
"RandomForest": RandomForestClassifier(
n_estimators=skrub.choose_int(20, 200, log=True)
)
}, name="regressor"
)
)
search = regressor.skb.make_randomized_search(scoring="roc_auc", fitted=True)Tuning with DataOps is not limited to estimators
'- choose_from([<Expr [\'col("grade").mean()\'] at 0x783825B3A3D0>, <Expr [\'col("grade").count()\'] at 0x78382C9A0090>]): [<Expr [\'col("grade").mean()\'] at 0x783825B3A3D0>, <Expr [\'col("grade").count()\'] at 0x78382C9A0090>]\n'Run hyperparameter search
Observe the impact of the hyperparameters
Data Ops provide a built-in parallel coordinate plot.
More information about the Data Ops
- Skrub example gallery
- Tutorial on timeseries forecasting at Euroscipy 2025
- Skrub User guide
- A Kaggle notebook on addressing the Titanic survival challenge with Data Ops
Wrapping up


Getting involved
Do you want to learn more?
Follow skrub on:
Star skrub on GitHub, or contribute directly:
tl;dw
skrub provides
- interactive data exploration
- automated pre-processing of pandas and polars dataframes
- powerful feature engineering
- DataOps, plans, hyperparameter tuning, (almost) no leakage
https://skrub-data.org/skrub-materials/