id,code,name,slug
15,76,Occitanie,occitanie
16,84,Auvergne-Rhône-Alpes,"auvergne rhone alpes"
17,93,"Provence-Alpes-Côte d'Azur","provence alpes cote dazur"
18,94,Corse,corse
19,COM,"Collectivités d'Outre-Mer","collectivites doutre mer"Skrub
Machine learning with dataframes
Inria P16
2025-11-21
whoami
I am a research engineer at Inria as part of the SODA team and the P16 project, and I am the lead developer of skrub
![]()
I’m Italian, but I don’t drink coffee, wine, and I like pizza with fries
![]()
I did my PhD in Côte d’Azur, and I moved away because it was too sunny and I don’t like the sea
![]()
Building a predictive pipeline with skrub
Premise and warning
Important
This lecture should give you an idea of possible problems you might run into, and how skrub can help you deal with (some of) them. The idea is giving you material you can refer back to if you encounter the same issue later on.
Skrub compatibility
- Skrub is fully compatible with pandas and polars
- Skrub transformers are fully compatible with scikit-learn
An example pipeline
- Gather some data
- Explore the data
- Preprocess the data
- Perform feature engineering
- Build a scikit-learn pipeline
- ???
- Profit?
Gathering the data: reading files, parsing dtypes
What happens if pandas parses this?
What happens if pandas parses this?
How about now?
Department code;Department name;Town code;Town name;Registered;Abstentions;Null;Choice A;Choice B
ZZ;FRANCAIS DE L'ETRANGER;8;Europe du Sud, Turquie, Israël;109763;84466;292;9299;15706
ZZ;FRANCAIS DE L'ETRANGER;9;Afrique Nord-Ouest;98997;59887;321;22116;16673
ZZ;FRANCAIS DE L'ETRANGER;10;Afrique Centre, Sud et Est;89859;46782;566;17008;25503
ZZ;FRANCAIS DE L'ETRANGER;11;Europe de l'est, Asie, Océanie;80061;42911;488;13975;22687How about now?
| Department code;Department name;Town code;Town name;Registered;Abstentions;Null;Choice A;Choice B | ||
|---|---|---|
| ZZ;FRANCAIS DE L'ETRANGER;8;Europe du Sud | Turquie | Israël;109763;84466;292;9299;15706 |
| ZZ;FRANCAIS DE L'ETRANGER;9;Afrique Nord-Ouest;98997;59887;321;22116;16673 | NaN | NaN |
| ZZ;FRANCAIS DE L'ETRANGER;10;Afrique Centre | Sud et Est;89859;46782;566;17008;25503 | NaN |
| ZZ;FRANCAIS DE L'ETRANGER;11;Europe de l'est | Asie | Océanie;80061;42911;488;13975;22687 |
Parsing CSV files is difficult!
- CSV files are plain text files and do not store any info besides what you can see.
- Any information about dtypes is lost.
- It is very hard to automate discovering the separator (ambiguity is unavoidable).
- Files cannot be seeked: to get to a line, all the previous lines must be read first.
- Plain text is uncompressed and can take up a very large amount of space.
- Adding metadata is difficult and may make parsing harder.
CSV vs Parquet
- Apache Parquet is a binary data storage format that addresses various issues with storing data.
- Dtypes are well defined and fixed.
- No separators to deal with: each column is defined in a schema when the parquet file is created.
- Additional metadata can be stored to provide context to the data.
- Parquet files are seekable: no need to read through the entire thing to get to a specific line.
- Parquet files are compressed, reducing the size by multiple times (depending on data)
- Parquet files are not human-readable. CSV files are, but are you really going to manually go through a few million lines of text?
Pandas vs Polars with Parquet
- Pandas requires additional dependencies to read Parquet files (
pyarroworfastparquet). - Polars supports Parquet files natively and includes additional features that make use of the format.
Exploring the data
Exploratory analysis
Before starting any kind of training, it’s important to explore the data:
- How big is the dataset (on disk, in rows/columns etc.)?
- What kind of datatypes are we dealing with? Anything that needs more attention?
- What is the distribution of values in a column?
- Are there null values? In what columns?
- Are there columns that may leak information to my model?
- Are there columns that look problematic? (this takes experience)
This is not an exhaustive list!
Exploring the data with Pandas: parsing
| gender | department | department_name | division | assignment_category | employee_position_title | date_first_hired | year_first_hired | |
|---|---|---|---|---|---|---|---|---|
| 0 | F | POL | Department of Police | MSB Information Mgmt and Tech Division Records... | Fulltime-Regular | Office Services Coordinator | 09/22/1986 | 1986 |
| 1 | M | POL | Department of Police | ISB Major Crimes Division Fugitive Section | Fulltime-Regular | Master Police Officer | 09/12/1988 | 1988 |
| 2 | F | HHS | Department of Health and Human Services | Adult Protective and Case Management Services | Fulltime-Regular | Social Worker IV | 11/19/1989 | 1989 |
| 3 | M | COR | Correction and Rehabilitation | PRRS Facility and Security | Fulltime-Regular | Resident Supervisor II | 05/05/2014 | 2014 |
| 4 | M | HCA | Department of Housing and Community Affairs | Affordable Housing Programs | Fulltime-Regular | Planning Specialist III | 03/05/2007 | 2007 |
Exploring the data: describe
| gender | department | department_name | division | assignment_category | employee_position_title | date_first_hired | year_first_hired | |
|---|---|---|---|---|---|---|---|---|
| count | 9211 | 9228 | 9228 | 9228 | 9228 | 9228 | 9228 | 9228.000000 |
| unique | 2 | 37 | 37 | 694 | 2 | 443 | 2264 | NaN |
| top | M | POL | Department of Police | School Health Services | Fulltime-Regular | Bus Operator | 12/12/2016 | NaN |
| freq | 5481 | 1844 | 1844 | 300 | 8394 | 638 | 87 | NaN |
| mean | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 2003.597529 |
| std | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 9.327078 |
| min | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 1965.000000 |
| 25% | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 1998.000000 |
| 50% | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 2005.000000 |
| 75% | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 2012.000000 |
| max | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 2016.000000 |
Exploring the data: column info
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 9228 entries, 0 to 9227
Data columns (total 8 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 gender 9211 non-null object
1 department 9228 non-null object
2 department_name 9228 non-null object
3 division 9228 non-null object
4 assignment_category 9228 non-null object
5 employee_position_title 9228 non-null object
6 date_first_hired 9228 non-null object
7 year_first_hired 9228 non-null int64
dtypes: int64(1), object(7)
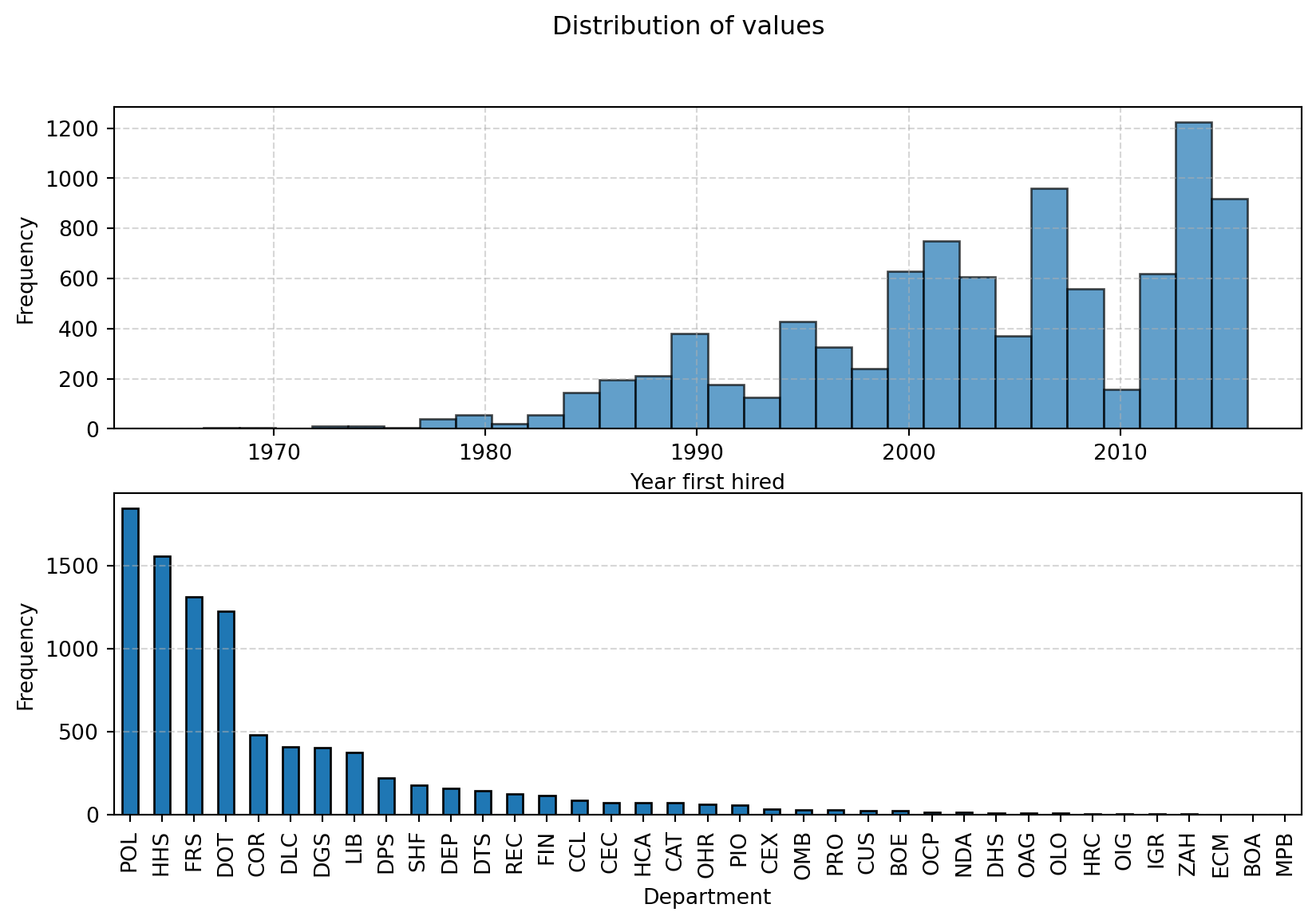
memory usage: 576.9+ KBExploring the data: distributions
import matplotlib.pyplot as plt
from pprint import pprint
# Plot the distribution of the numerical values using a histogram
fig, axs = plt.subplots(2,1, figsize=(10, 6))
ax1, ax2 = axs
ax1.hist(df['year_first_hired'], bins=30, edgecolor='black', alpha=0.7)
ax1.set_xlabel('Year first hired')
ax1.set_ylabel('Frequency')
ax1.grid(True, linestyle='--', alpha=0.5)
# Count the frequency of each category
category_counts = df['department'].value_counts()
# Create a bar plot
category_counts.plot(kind='bar', edgecolor='black', ax=ax2)
# Add labels and title
ax2.set_xlabel('Department')
ax2.set_ylabel('Frequency')
ax2.grid(True, linestyle='--', axis='y', alpha=0.5) # Add grid lines for y-axis
fig.suptitle("Distribution of values")
# Show the plot
plt.show()Exploring the data: distributions
Exploring the data with skrub
Main features:
- Obtain high-level statistics about the data
- Explore the distribution of values and find outliers
- Discover highly correlated columns
- Export and share the report as an
htmlfile
Exploring the data with skrub
Things to notice:
- Shape of the dataframe (rows, columns)
- Dtype of the columns (string, numerical, datetime)
- Were dtypes converted properly?
- Missing values
- Are there columns with many missing values?
- Distribution of values
- Are there columns with high cardinality?
- Are there outliers?
- Are there columns with imbalanced distributions?
- Column associations
- Are there correlated columns?
Pre-processing
Data cleaning with pandas/polars: setup
import pandas as pd
import numpy as np
data = {
"Int": [2, 3, 2], # Multiple unique values
"Const str": ["x", "x", "x"], # Single unique value
"Str": ["foo", "bar", "baz"], # Multiple unique values
"All nan": [np.nan, np.nan, np.nan], # All missing values
"All empty": ["", "", ""], # All empty strings
"Date": ["01 Jan 2023", "02 Jan 2023", "03 Jan 2023"],
}
df_pd = pd.DataFrame(data)
display(df_pd)| Int | Const str | Str | All nan | All empty | Date | |
|---|---|---|---|---|---|---|
| 0 | 2 | x | foo | NaN | 01 Jan 2023 | |
| 1 | 3 | x | bar | NaN | 02 Jan 2023 | |
| 2 | 2 | x | baz | NaN | 03 Jan 2023 |
import polars as pl
import numpy as np
data = {
"Int": [2, 3, 2], # Multiple unique values
"Const str": ["x", "x", "x"], # Single unique value
"Str": ["foo", "bar", "baz"], # Multiple unique values
"All nan": [np.nan, np.nan, np.nan], # All missing values
"All empty": ["", "", ""], # All empty strings
"Date": ["01 Jan 2023", "02 Jan 2023", "03 Jan 2023"],
}
df_pl = pl.DataFrame(data)
display(df_pl)| Int | Const str | Str | All nan | All empty | Date |
|---|---|---|---|---|---|
| i64 | str | str | f64 | str | str |
| 2 | "x" | "foo" | NaN | "" | "01 Jan 2023" |
| 3 | "x" | "bar" | NaN | "" | "02 Jan 2023" |
| 2 | "x" | "baz" | NaN | "" | "03 Jan 2023" |
Nulls, datetimes, constant columns with pandas/polars
# Parse the datetime strings with a specific format
df_pd['Date'] = pd.to_datetime(df_pd['Date'], format='%d %b %Y')
# Drop columns with only a single unique value
df_pd_cleaned = df_pd.loc[:, df_pd.nunique(dropna=True) > 1]
# Function to drop columns with only missing values or empty strings
def drop_empty_columns(df):
# Drop columns with only missing values
df_cleaned = df.dropna(axis=1, how='all')
# Drop columns with only empty strings
empty_string_cols = df_cleaned.columns[df_cleaned.eq('').all()]
df_cleaned = df_cleaned.drop(columns=empty_string_cols)
return df_cleaned
# Apply the function to the DataFrame
df_pd_cleaned = drop_empty_columns(df_pd_cleaned)# Parse the datetime strings with a specific format
df_pl = df_pl.with_columns([
pl.col("Date").str.strptime(pl.Date, "%d %b %Y", strict=False).alias("Date")
])
# Drop columns with only a single unique value
df_pl_cleaned = df_pl.select([
col for col in df_pl.columns if df_pl[col].n_unique() > 1
])
# Import selectors for dtype selection
import polars.selectors as cs
# Drop columns with only missing values or only empty strings
def drop_empty_columns(df):
all_nan = df.select(
[
col for col in df.select(cs.numeric()).columns if
df [col].is_nan().all()
]
).columns
all_empty = df.select(
[
col for col in df.select(cs.string()).columns if
(df[col].str.strip_chars().str.len_chars()==0).all()
]
).columns
to_drop = all_nan + all_empty
return df.drop(to_drop)
df_pl_cleaned = drop_empty_columns(df_pl_cleaned)Data cleaning with skrub.Cleaner
Cleaning employee_salaries
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 9228 entries, 0 to 9227
Data columns (total 8 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 gender 9211 non-null object
1 department 9228 non-null object
2 department_name 9228 non-null object
3 division 9228 non-null object
4 assignment_category 9228 non-null object
5 employee_position_title 9228 non-null object
6 date_first_hired 9228 non-null object
7 year_first_hired 9228 non-null int64
dtypes: int64(1), object(7)
memory usage: 576.9+ KBCleaning employee_salaries
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 9228 entries, 0 to 9227
Data columns (total 8 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 gender 9211 non-null object
1 department 9228 non-null object
2 department_name 9228 non-null object
3 division 9228 non-null object
4 assignment_category 9228 non-null object
5 employee_position_title 9228 non-null object
6 date_first_hired 9228 non-null datetime64[ns]
7 year_first_hired 9228 non-null int64
dtypes: datetime64[ns](1), int64(1), object(6)
memory usage: 576.9+ KBThe Cleaner…
- Automatically parses numerical values, even if they have string as dtype
- Tries to parse datetimes, or takes a format from the user
- Allows to drop uninformative columns:
- All nulls
- A single value
- All unique values (careful!)
Feature engineering
Encoding datetime features with pandas/polars
import pandas as pd
data = {
'date': ['2023-01-01 12:34:56', '2023-02-15 08:45:23', '2023-03-20 18:12:45'],
'value': [10, 20, 30]
}
df_pd = pd.DataFrame(data)
datetime_column = "date"
df_pd[datetime_column] = pd.to_datetime(df_pd[datetime_column], errors='coerce')
df_pd['year'] = df_pd[datetime_column].dt.year
df_pd['month'] = df_pd[datetime_column].dt.month
df_pd['day'] = df_pd[datetime_column].dt.day
df_pd['hour'] = df_pd[datetime_column].dt.hour
df_pd['minute'] = df_pd[datetime_column].dt.minute
df_pd['second'] = df_pd[datetime_column].dt.secondimport polars as pl
data = {
'date': ['2023-01-01 12:34:56', '2023-02-15 08:45:23', '2023-03-20 18:12:45'],
'value': [10, 20, 30]
}
df_pl = pl.DataFrame(data)
df_pl = df_pl.with_columns(date=pl.col("date").str.to_datetime())
df_pl = df_pl.with_columns(
year=pl.col("date").dt.year(),
month=pl.col("date").dt.month(),
day=pl.col("date").dt.day(),
hour=pl.col("date").dt.hour(),
minute=pl.col("date").dt.minute(),
second=pl.col("date").dt.second(),
)Adding periodic features with pandas/polars
Encoding datetime features with skrub.DatetimeEncoder
shape: (3, 7)
┌───────────┬────────────┬──────────┬───────────┬─────────────┬─────────────┬────────────────────┐
│ date_year ┆ date_month ┆ date_day ┆ date_hour ┆ date_minute ┆ date_second ┆ date_total_seconds │
│ --- ┆ --- ┆ --- ┆ --- ┆ --- ┆ --- ┆ --- │
│ f32 ┆ f32 ┆ f32 ┆ f32 ┆ f32 ┆ f32 ┆ f32 │
╞═══════════╪════════════╪══════════╪═══════════╪═════════════╪═════════════╪════════════════════╡
│ 2023.0 ┆ 1.0 ┆ 1.0 ┆ 12.0 ┆ 34.0 ┆ 56.0 ┆ 1.6726e9 │
│ 2023.0 ┆ 2.0 ┆ 15.0 ┆ 8.0 ┆ 45.0 ┆ 23.0 ┆ 1.6765e9 │
│ 2023.0 ┆ 3.0 ┆ 20.0 ┆ 18.0 ┆ 12.0 ┆ 45.0 ┆ 1.6793e9 │
└───────────┴────────────┴──────────┴───────────┴─────────────┴─────────────┴────────────────────┘Encoding datetime features skrub.DatetimeEncoder
shape: (3, 8)
┌───────────┬────────────┬────────────┬────────────┬───────────┬───────────┬───────────┬───────────┐
│ date_year ┆ date_total ┆ date_month ┆ date_month ┆ date_day_ ┆ date_day_ ┆ date_hour ┆ date_hour │
│ --- ┆ _seconds ┆ _circular_ ┆ _circular_ ┆ circular_ ┆ circular_ ┆ _circular ┆ _circular │
│ f32 ┆ --- ┆ 0 ┆ 1 ┆ 0 ┆ 1 ┆ _0 ┆ _1 │
│ ┆ f32 ┆ --- ┆ --- ┆ --- ┆ --- ┆ --- ┆ --- │
│ ┆ ┆ f64 ┆ f64 ┆ f64 ┆ f64 ┆ f64 ┆ f64 │
╞═══════════╪════════════╪════════════╪════════════╪═══════════╪═══════════╪═══════════╪═══════════╡
│ 2023.0 ┆ 1.6726e9 ┆ 0.5 ┆ 0.866025 ┆ 0.207912 ┆ 0.978148 ┆ 1.2246e-1 ┆ -1.0 │
│ ┆ ┆ ┆ ┆ ┆ ┆ 6 ┆ │
│ 2023.0 ┆ 1.6765e9 ┆ 0.866025 ┆ 0.5 ┆ 1.2246e-1 ┆ -1.0 ┆ 0.866025 ┆ -0.5 │
│ ┆ ┆ ┆ ┆ 6 ┆ ┆ ┆ │
│ 2023.0 ┆ 1.6793e9 ┆ 1.0 ┆ 6.1232e-17 ┆ -0.866025 ┆ -0.5 ┆ -1.0 ┆ -1.8370e- │
│ ┆ ┆ ┆ ┆ ┆ ┆ ┆ 16 │
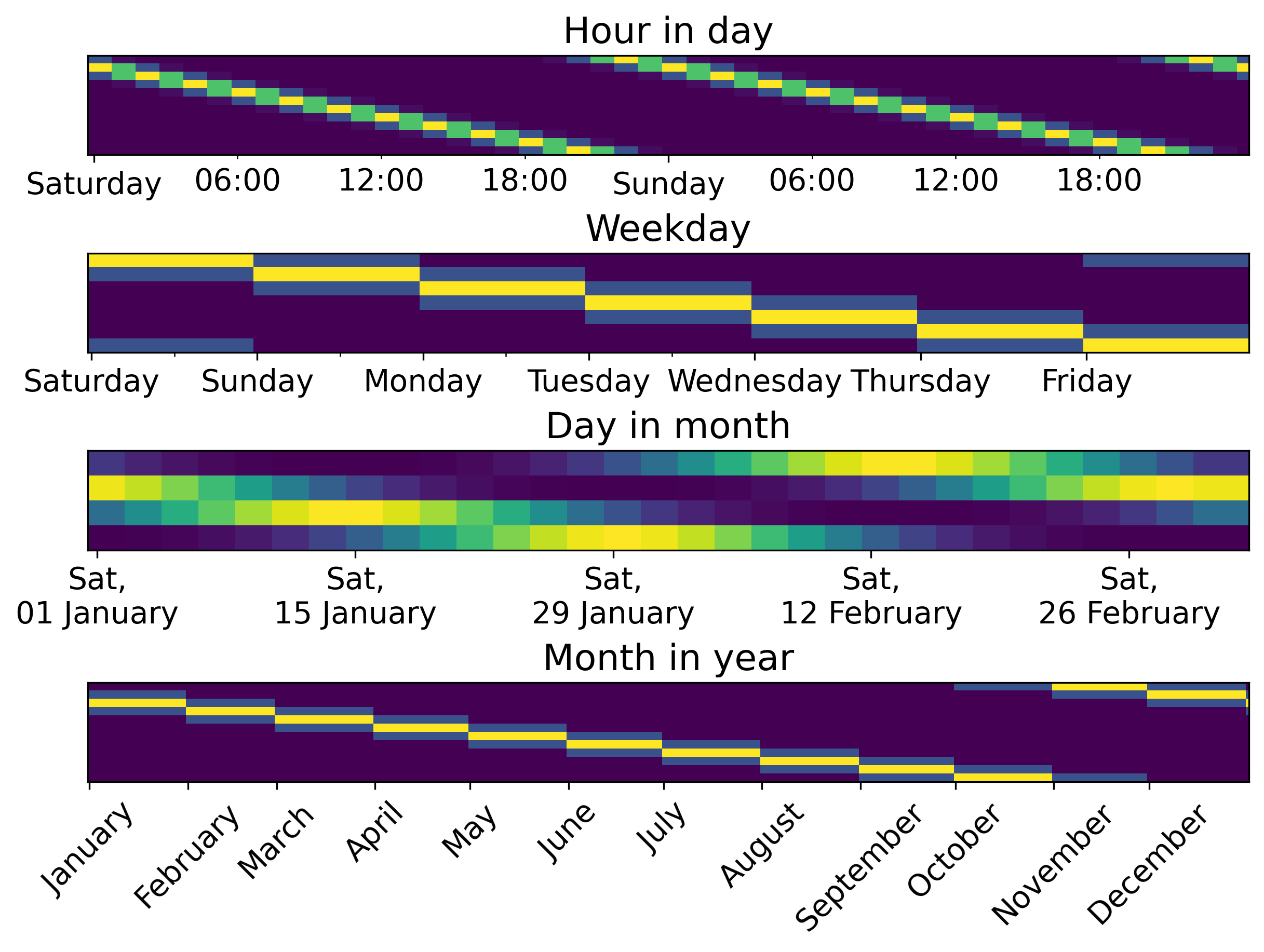
└───────────┴────────────┴────────────┴────────────┴───────────┴───────────┴───────────┴───────────┘What periodic features look like
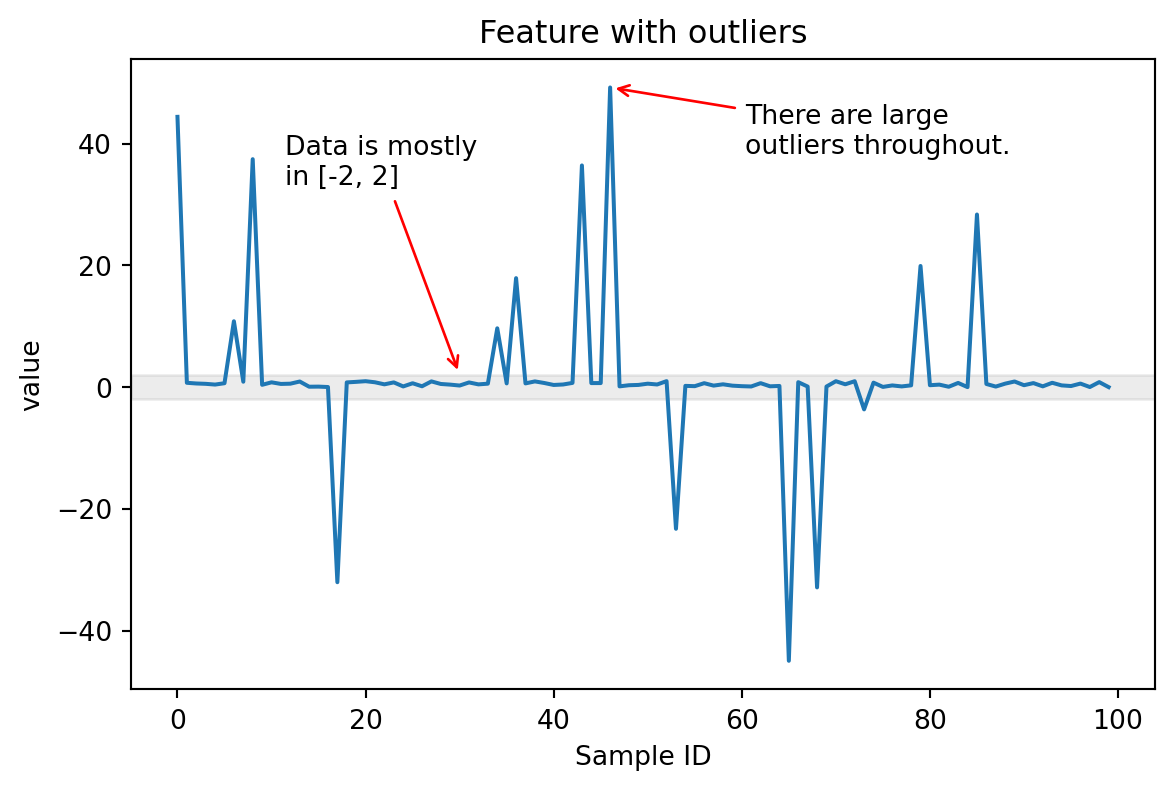
Scaling numerical features: outliers!
Comparing different scalers in presence of outliers
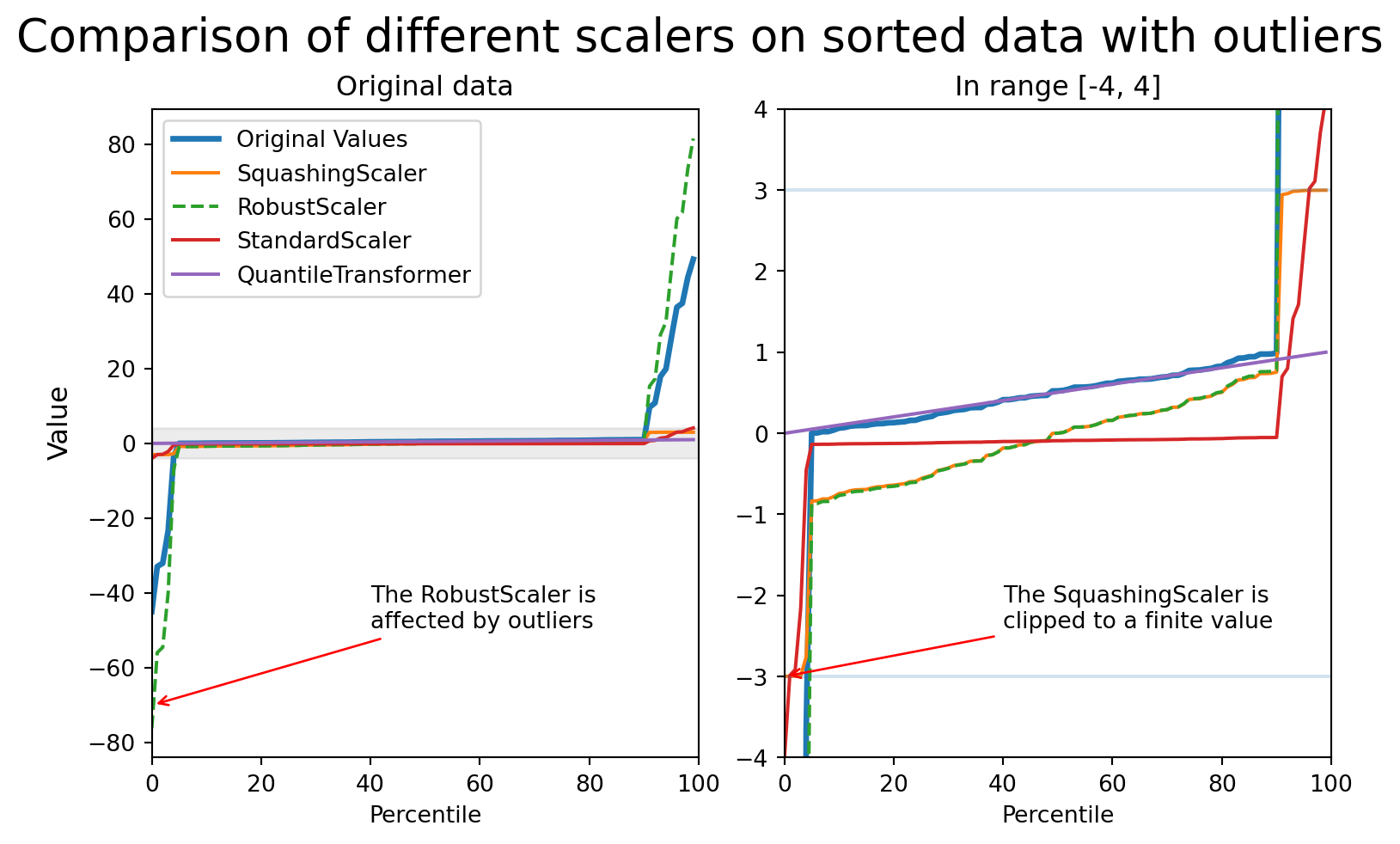
Dealing with categorical features
- “Categorical features” include all features that belong to “discrete categories”.
- For example, actual categories (grades, clothing sizes, countries…).
- Text is also categorical! Any unique string is a category.
- There may be many unique categories.
How do you deal with that?
Basic strategies: OneHotEncoder
- The
OneHotEncodercreates an indicator column for each unique value in a column. - Fine if you have few categories, problematic if you have many.
from sklearn.preprocessing import OneHotEncoder
df = fetch_employee_salaries().X
enc = OneHotEncoder(sparse_output=False).fit_transform(X=df[["gender"]])
print("The number of unique values in column `gender` is: ", df[["gender"]].nunique().item())
print("The shape of the OneHotEncoding of column `gender` is: ", enc.shape)The number of unique values in column `gender` is: 2
The shape of the OneHotEncoding of column `gender` is: (9228, 3)Looks fine!
Basic strategies: OneHotEncoder
What about a different column?
The number of unique values in column `division` is: 694
The shape of the OneHotEncoding of column `division` is: (9228, 694)Warning
That’s ~700 features that are mostly 0’s.
Remember that more features mean:
- Overfitting
- Worse performance
- Increased training time and resources needed
- Worse interpretability
Basic strategies: OrdinalEncoder
The number of unique values in column `division` is: 694
The shape of the OneHotEncoding of column `division` is: (9228, 1)Now we’re keeping the number of dimensions down, but we are introducing an ordering that may not make any sense in reality.
Encoding categorical (string/text) features
Categorical features have a “cardinality”: the number of unique values
- Low cardinality features:
OneHotEncoder.- The
OneHotEncoderproduces sparse matrices. Dataframes are dense.
- The
- High cardinality features (>40 unique values):
- Using
OneHotEncodergenerates too many (dense) features. - Use encoders with a fixed number of output features.
- Using
- Latent Semantic Analysis with:
skrub.StringEncoder- Apply tf-idf to the ngrams in the column, followed by SVD.
- Robust, quick, fixed number of output features regardless of # of unique values.
Encoding categorical (string/text) features
- Textual features:
skrub.TextEncoderand pretrained models from HuggingFace Hub.- The
TextEncoderneeds PyTorch: very heavy dependency. - Models are large and take time to download.
- Encoding is much slower.
- However, performance can be much better depending on the dataset.
- The
Deeper dive in this post.
Building pipelines
Encoding all the features: TableVectorizer
- Apply the
Cleanerto all columns - Split columns by dtype and # of unique values
- Encode each column separately
Encoding all the features: TableVectorizer

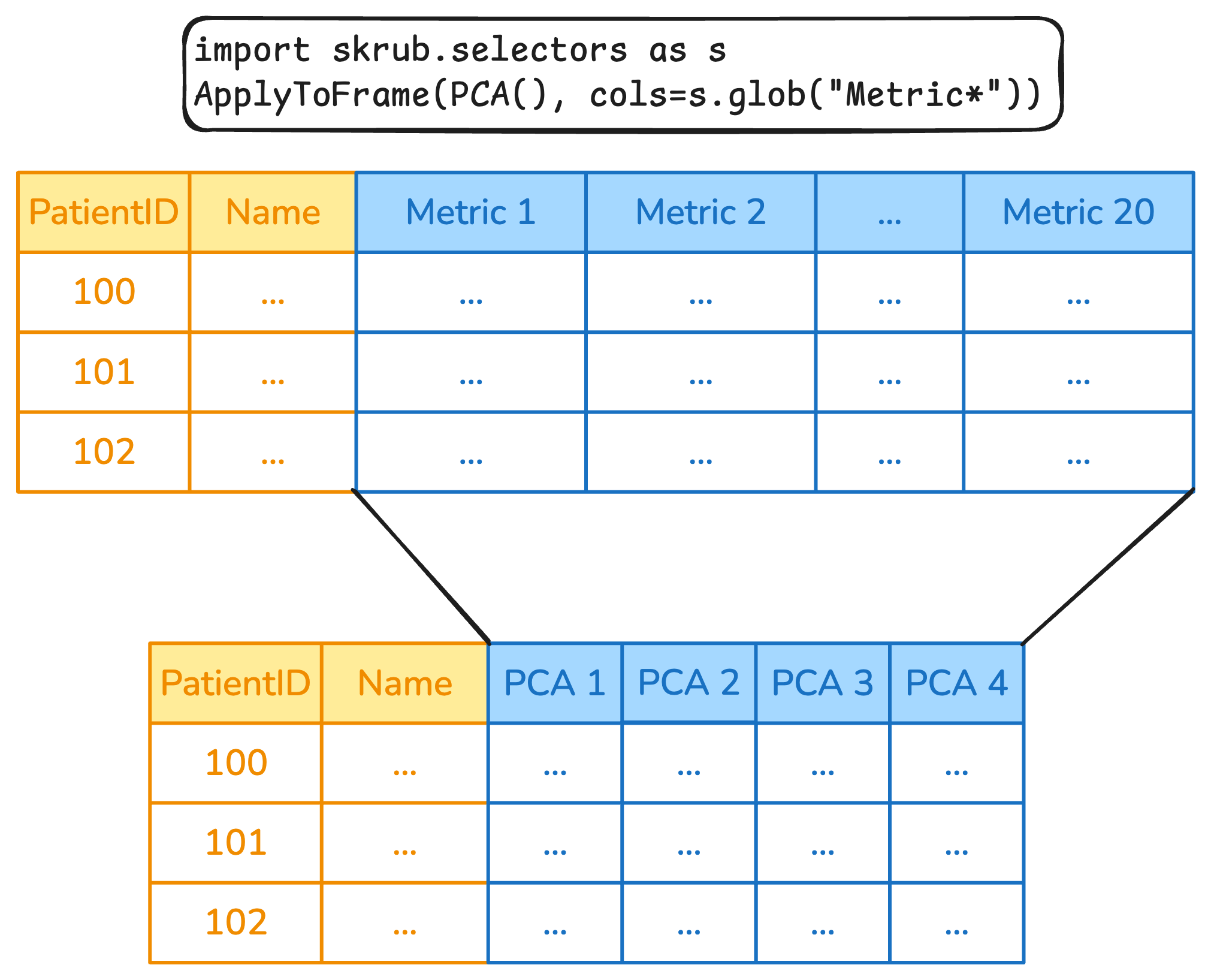
Fine-grained column transformations
- Various skrub transformers can accept a
colsparameter that lets you select what columns should be modified. SelectCols,DropCols,ApplyToCols,ApplyToFrame, and.skb.apply()can takecolsas a parameter.- Complex selection rules can be implemented with
skrub.selectors.
Why is column selection important?
- There is an index I don’t want to touch
- Column names give me information about the content
- I want to transform only datetimes (or numbers, or strings…)
- I need to combine conditions (column has nulls AND column is numeric)
Selecting columns with the skrub selectors
| height_mm | width_mm | kind | ID | |
|---|---|---|---|---|
| 0 | 297.0 | 210.0 | A4 | 4 |
| 1 | 420.0 | 297.0 | A3 | 3 |
Selecting columns with the skrub selectors
Select all columns based on the name:
Select columns based on dtypes, and invert selections:
More info in the selectors user guide.
Column transformations with ApplyToCols and ApplyToFrame
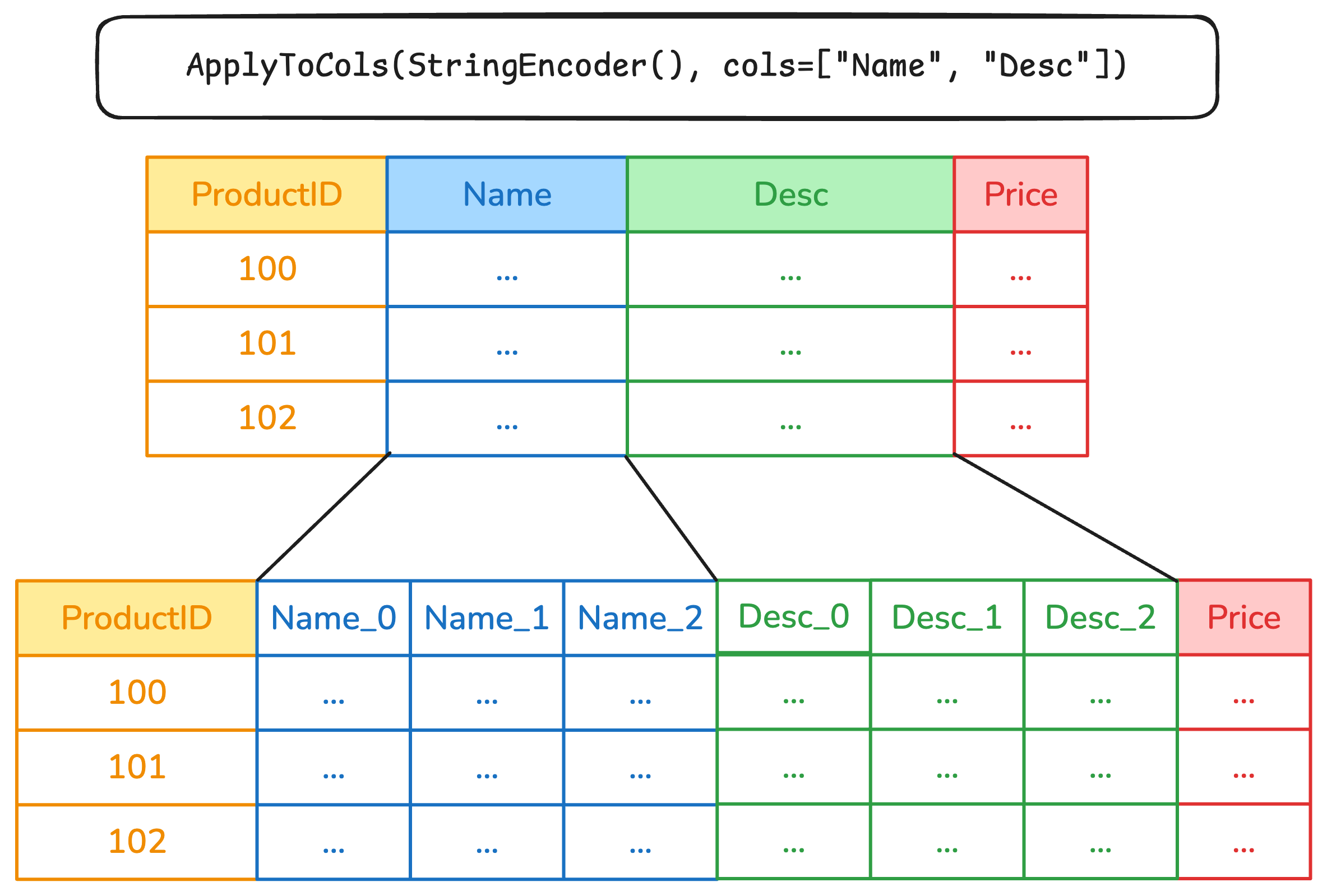
Column transformations with ApplyToCols and ApplyToFrame
Replacing ColumnTransformer with ApplyToCols
import pandas as pd
from sklearn.compose import make_column_selector as selector
from sklearn.compose import make_column_transformer
from sklearn.preprocessing import StandardScaler, OneHotEncoder
df = pd.DataFrame({"text": ["foo", "bar", "baz"], "number": [1, 2, 3]})
categorical_columns = selector(dtype_include=object)(df)
numerical_columns = selector(dtype_exclude=object)(df)
ct = make_column_transformer(
(StandardScaler(),
numerical_columns),
(OneHotEncoder(handle_unknown="ignore"),
categorical_columns))
transformed = ct.fit_transform(df)
transformedarray([[-1.22474487, 0. , 0. , 1. ],
[ 0. , 1. , 0. , 0. ],
[ 1.22474487, 0. , 1. , 0. ]])Replacing ColumnTransformer with ApplyToCols
import skrub.selectors as s
from sklearn.pipeline import make_pipeline
from skrub import ApplyToCols
numeric = ApplyToCols(StandardScaler(), cols=s.numeric())
string = ApplyToCols(OneHotEncoder(sparse_output=False), cols=s.string())
transformed = make_pipeline(numeric, string).fit_transform(df)
transformed| text_bar | text_baz | text_foo | number | |
|---|---|---|---|---|
| 0 | 0.0 | 0.0 | 1.0 | -1.224745 |
| 1 | 1.0 | 0.0 | 0.0 | 0.000000 |
| 2 | 0.0 | 1.0 | 0.0 | 1.224745 |
Let’s build a predictive pipeline
Let’s build a predictive pipeline
Let’s build a predictive pipeline
from sklearn.linear_model import Ridge
from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import StandardScaler, OneHotEncoder
from sklearn.impute import SimpleImputer
from sklearn.compose import make_column_selector as selector
from sklearn.compose import make_column_transformer
categorical_columns = selector(dtype_include=object)(employees)
numerical_columns = selector(dtype_exclude=object)(employees)
ct = make_column_transformer(
(StandardScaler(),
numerical_columns),
(OneHotEncoder(handle_unknown="ignore"),
categorical_columns))
model = make_pipeline(ct, SimpleImputer(), Ridge())Now with tabular_pipeline
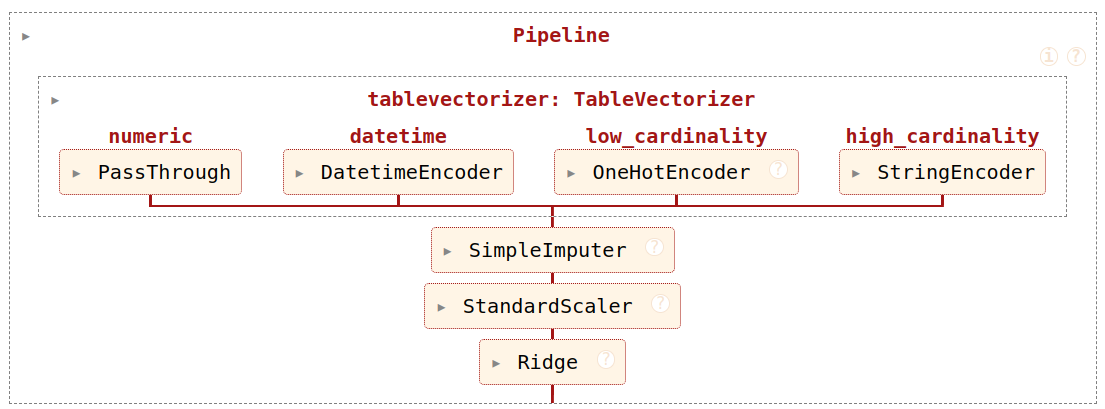

We now have a pipeline!
- Gather some data
- Explore the data
TableReport
- Pre-process the data
Cleaner…
- Perform feature engineering
TableVectorizer,SquashingScaler,TextEncoder,StringEncoder…
- Build a scikit-learn pipeline
tabular_pipeline,sklearn.pipeline.make_pipeline…
- ???
- Profit 📈
Skrub Data Ops
Disclaimer
Warning
This section covers more advanced topics that are more representative of what happens in real scenarios, and how skrub can help addressing some of them.
What if…
- Your data is spread over multiple tables?
- You want to avoid data leakage?
- You want to tune more than just the hyperparameters of your model?
- You want to guarantee that your pipeline is replayed exactly on new data?
When a normal pipeline is not enough…
… the skrub DataOps come to the rescue 🚒
DataOps…
- Extend the
scikit-learnmachinery to complex multi-table operations. - Take care of data leakage.
- Track all operations with a computational graph (a Data Ops plan).
- Allow tuning any operation in the Data Ops plan.
- Can be persisted and shared easily.
How do DataOps work, though?
DataOps wrap around user operations, where user operations are:
- any dataframe operation (e.g., merge, group by, aggregate etc.).
- scikit-learn estimators (a Random Forest, RidgeCV etc.).
- custom user code (load data from a path, fetch from an URL etc.).
Important
DataOps record user operations, so that they can later be replayed in the same order and with the same arguments on unseen data.
Looking at the computational graph
Each node:
- Shows a preview of the data resulting from the operation
- Reports the location in the code where the code is defined
- Shows the run time of the node
- Offline and in HTML, no need for a running kernel
Tuning in scikit-learn can be complex
pipe = Pipeline([("dim_reduction", PCA()), ("regressor", Ridge())])
grid = [
{
"dim_reduction": [PCA()],
"dim_reduction__n_components": [10, 20, 30],
"regressor": [Ridge()],
"regressor__alpha": loguniform(0.1, 10.0),
},
{
"dim_reduction": [SelectKBest()],
"dim_reduction__k": [10, 20, 30],
"regressor": [Ridge()],
"regressor__alpha": loguniform(0.1, 10.0),
},
{
"dim_reduction": [PCA()],
"dim_reduction__n_components": [10, 20, 30],
"regressor": [RandomForestRegressor()],
"regressor__n_estimators": loguniform(20, 200),
},
{
"dim_reduction": [SelectKBest()],
"dim_reduction__k": [10, 20, 30],
"regressor": [RandomForestRegressor()],
"regressor__n_estimators": loguniform(20, 200),
},
]
model = RandomizedSearchCV(pipe, grid)Tuning with DataOps is simple!
dim_reduction = X.skb.apply(
skrub.choose_from(
{
"PCA": PCA(n_components=skrub.choose_int(10, 30)),
"SelectKBest": SelectKBest(k=skrub.choose_int(10, 30))
}, name="dim_reduction"
)
)
regressor = dim_reduction.skb.apply(
skrub.choose_from(
{
"Ridge": Ridge(alpha=skrub.choose_float(0.1, 10.0, log=True)),
"RandomForest": RandomForestRegressor(
n_estimators=skrub.choose_int(20, 200, log=True)
)
}, name="regressor"
)
)
search = regressor.skb.make_randomized_search(scoring="roc_auc", fitted=True)Run hyperparameter search
Hyperparameter tuning with choose_*
skrub implements four choose_* functions:
choose_from: select from the given list of optionschoose_int: select an integer within a rangechoose_float: select a float within a rangechoose_bool: select a booloptional: chooses whether to execute the given operation
Tuning with DataOps is not limited to estimators
"- choose_from(['count', 'mean', 'max']): ['count', 'mean', 'max']\n"'- choose_from([<Expr [\'col("grade").mean()\'] at 0x354194A50>, <Expr [\'col("grade").count()\'] at 0x354194B50>]): [<Expr [\'col("grade").mean()\'] at 0x354194A50>, <Expr [\'col("grade").count()\'] at 0x354194B50>]\n'How does tuning affect the results?
Data Ops provide a built-in parallel coordinate plot.
Exporting the plan in a learner
The Learner is a stand-alone object that works like a scikit-learn estimator that takes a dictionary as input rather than just X and y.
Then, the learner can be pickled …
More information about Data Ops
- Skrub example gallery
- Tutorial on timeseries forecasting at Euroscipy 2025
- Skrub User guide
- A Kaggle notebook on addressing the Titanic survival challenge with Data Ops
Wrapping up


Getting involved
Do you want to learn more?
Follow skrub on:
Star skrub on GitHub, or contribute directly:
tl;dw
skrub provides
- interactive data exploration
- automated pre-processing of pandas and polars dataframes
- powerful feature engineering
- DataOps, plans, hyperparameter tuning, (almost) no leakage
https://skrub-data.org/skrub-materials/


