MinHashEncoder#
- class skrub.MinHashEncoder(*, n_components=30, ngram_range=(2, 4), hashing='fast', minmax_hash=False, n_jobs=None)[source]#
Encode string categorical features by applying the MinHash method to n-gram decompositions of strings.
Note
MinHashEncoderis a type of single-column transformer. Unlike most scikit-learn estimators, itsfit,transformandfit_transformmethods expect a single column (a pandas or polars Series) rather than a full dataframe. To apply this transformer to one or more columns in a dataframe, use it as a parameter in askrub.TableVectorizerorsklearn.compose.ColumnTransformer. In theColumnTransformer, pass a single column:make_column_transformer((MinHashEncoder(), 'col_name_1'), (MinHashEncoder(), 'col_name_2'))instead ofmake_column_transformer((MinHashEncoder(), ['col_name_1', 'col_name_2'])).The principle is as follows:
A string is viewed as a succession of numbers (the ASCII or UTF8 representation of its elements).
The string is then decomposed into a set of n-grams, i.e. n-dimensional vectors of integers.
A hashing function is used to assign an integer to each n-gram. The minimum of the hashes over all n-grams is used in the encoding.
This process is repeated with N hashing functions to form N-dimensional encodings.
Maxhash encodings can be computed similarly by taking the maximum hash instead. With this procedure, strings that share many n-grams have a greater probability of having the same encoding value. These encodings thus capture morphological similarities between strings.
- Parameters:
- n_components
int, default=30 The number of dimension of encoded strings. Numbers around 300 tend to lead to good prediction performance, but with more computational cost.
- ngram_range2-tuple of
int, default=(2, 4) The lower and upper boundaries of the range of n-values for different n-grams used in the string similarity. All values of n such that
min_n <= n <= max_nwill be used.- hashing{‘fast’, ‘murmur’}, default=’fast’
Hashing function. fast is faster than murmur but might have some concern with its entropy.
- minmax_hash
bool, default=False If True, returns the min and max hashes concatenated.
- n_jobs
int, optional The number of jobs to run in parallel. The hash computations for all unique elements are parallelized. None means 1 unless in a joblib.parallel_backend. -1 means using all processors. See n_jobs for more details.
- n_components
- Attributes:
See also
GapEncoderEncodes dirty categories (strings) by constructing latent topics with continuous encoding.
SimilarityEncoderEncode string columns as a numeric array with n-gram string similarity.
StringEncoderFast n-gram encoding of string columns.
deduplicateDeduplicate data by hierarchically clustering similar strings.
References
For a detailed description of the method, see Encoding high-cardinality string categorical variables by Cerda, Varoquaux (2019).
Examples
>>> import pandas as pd >>> from skrub import MinHashEncoder >>> enc = MinHashEncoder(n_components=5)
Let’s encode the following non-normalized data:
>>> X = pd.Series(['paris, FR', 'Paris', 'London, UK', 'London'], name='city') >>> enc.fit(X) MinHashEncoder(n_components=5)
The encoded data with 5 components are:
>>> enc.transform(X) city_0 city_1 city_2 city_3 city_4 0 -1.783375e+09 -1.588270e+09 -1.663592e+09 -1.819887e+09 -1.962594e+09 1 -8.480470e+08 -1.766579e+09 -1.558912e+09 -1.485745e+09 -1.687299e+09 2 -1.975829e+09 -2.095000e+09 -1.596521e+09 -1.817594e+09 -2.095693e+09 3 -1.975829e+09 -2.095000e+09 -1.530721e+09 -1.459183e+09 -1.580988e+09
Methods
fit(X[, y])Fit the MinHashEncoder to X.
fit_transform(X[, y])Fit to data, then transform it.
get_feature_names_out([input_features])Return a list of features generated by the transformer.
get_params([deep])Get parameters for this estimator.
set_output(*[, transform])Set output container.
set_params(**params)Set the parameters of this estimator.
transform(X)Transform X using specified encoding scheme.
- fit(X, y=None)[source]#
Fit the MinHashEncoder to X.
In practice, just initializes a dictionary to store encodings to speed up computation.
- Parameters:
- Xarray_like, shape (n_samples, ) or (n_samples, n_columns)
The string data to encode. Only here for compatibility.
- y
None Unused, only here for compatibility.
- Returns:
MinHashEncoderThe fitted MinHashEncoder instance (self).
- fit_transform(X, y=None, **fit_params)[source]#
Fit to data, then transform it.
Fits transformer to X and y with optional parameters fit_params and returns a transformed version of X.
- Parameters:
- Xarray_like of shape (n_samples, n_features)
Input samples.
- yarray_like of shape (n_samples,) or (n_samples, n_outputs), default=None
Target values (None for unsupervised transformations).
- **fit_params
dict Additional fit parameters.
- Returns:
- get_feature_names_out(input_features=None)[source]#
Return a list of features generated by the transformer.
Each feature has format
{input_name}_{n_component}whereinput_nameis the name of the input column, or a default name for the encoder, andn_componentis the idx of the specific feature.
- set_output(*, transform=None)[source]#
Set output container.
See Introducing the set_output API for an example on how to use the API.
- Parameters:
- transform{“default”, “pandas”, “polars”}, default=None
Configure output of transform and fit_transform.
“default”: Default output format of a transformer
“pandas”: DataFrame output
“polars”: Polars output
None: Transform configuration is unchanged
Added in version 1.4: “polars” option was added.
- Returns:
- selfestimator instance
Estimator instance.
- set_params(**params)[source]#
Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as
Pipeline). The latter have parameters of the form<component>__<parameter>so that it’s possible to update each component of a nested object.- Parameters:
- **params
dict Estimator parameters.
- **params
- Returns:
- selfestimator instance
Estimator instance.
- transform(X)[source]#
Transform X using specified encoding scheme.
- Parameters:
- Xarray_like, shape (n_samples, ) or (n_samples, n_columns)
The string data to encode.
- Returns:
ndarrayof shape (n_samples, n_columns * n_components)Transformed input.
Gallery examples#
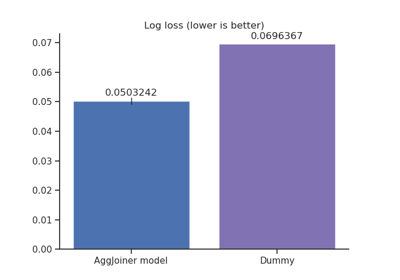
Encoding: from a dataframe to a numerical matrix for machine learning
Various string encoders: a sentiment analysis example
Multiples tables: building machine learning pipelines with DataOps