skrub.TableVectorizer#
Usage examples at the bottom of this page.
- class skrub.TableVectorizer(*, cardinality_threshold=40, low_cardinality=OneHotEncoder(drop='if_binary', dtype='float32', handle_unknown='ignore', sparse_output=False), high_cardinality=GapEncoder(n_components=30), numeric=PassThrough(), datetime=DatetimeEncoder(), specific_transformers=(), n_jobs=None)[source]#
Transform a dataframe to a numerical (vectorized) representation.
Applies a different transformation to each of several kinds of columns:
- numeric:
Floats, ints, and Booleans.
- datetime:
Datetimes and dates.
- low_cardinality:
String and categorical columns with a count of unique values smaller than a given threshold (40 by default). Category encoding schemes such as one-hot encoding, ordinal encoding etc. are typically appropriate for columns with few unique values.
- high_cardinality:
String and categorical columns with many unique values, such as free-form text. Such columns have so many distinct values that it is not possible to assign a distinct representation to each: the dimension would be too large and there would be too few examples of each category. Representations designed for text, such as topic modelling (GapEncoder) or locality-sensitive hashing (MinHash) are more appropriate.
Note
Transformations are applied independently on each column. A different transformer instance is used for each column separately; multivariate transformations are therefore not supported.
The transformer for each kind of column can be configured with the corresponding parameter.
A transformer can be a scikit-learn Transformer (an object providing the
fit,fit_transformandtransformmethods), a clone of which will be applied to each column separately. A transformer can also be the literal string"drop"to drop the corresponding columns (they will not appear in the output), or"passthrough"to leave them unchanged.Additionally, it is possible to specify transformers for specific columns, overriding the categorization described above. This is done by providing a list of pairs
(transformer, list_of_columns)as thespecific_transformersparameter.Note
The
specific_transformersparameter will be removed in a future version ofskrub, when better utilities for building complex pipelines are introduced.- Parameters:
- cardinality_threshold
int, default=40 String and categorical features with a number of unique values strictly smaller than this threshold are handled by the transformer
low_cardinality, the rest are handled by the transformerhigh_cardinality.- low_cardinalitytransformer, “passthrough” or “drop”, optional
The transformer for string or categorical columns with strictly fewer than
cardinality_thresholdunique values. The default is aOneHotEncoder.- high_cardinalitytransformer, “passthrough” or “drop”, optional
The transformer for string or categorical columns with at least
cardinality_thresholdunique values. The default is aGapEncoderwith 30 components (30 output columns for each input).- numerictransformer, “passthrough” or “drop”, optional
The transformer for numeric columns (floats, ints, booleans). The default is passthrough.
- datetimetransformer, “passthrough” or “drop”, optional
The transformer for date and datetime columns. The default is
DatetimeEncoder, which extracts features such as year, month, etc.- specific_transformers
listof (transformer,listof column names) pairs, optional Override the categories above for the given columns and force using the specified transformer. This disables any preprocessing usually done by the TableVectorizer; the columns are passed to the transformer without any modification. A column is not allowed to appear twice in
specific_transformers. Usingspecific_transformersprovides similar functionality to what is offered by scikit-learn’sColumnTransformer.- n_jobs
int, default=None Number of jobs to run in parallel.
Nonemeans 1 unless in a joblibparallel_backendcontext.-1means using all processors.
- cardinality_threshold
See also
tabular_learnerA function that accepts a scikit-learn estimator and creates a pipeline combining a
TableVectorizer, optional missing value imputation and the provided estimator.
Examples
>>> from skrub import TableVectorizer >>> import pandas as pd >>> df = pd.DataFrame({ ... 'A': ['one', 'two', 'two', 'three'], ... 'B': ['02/02/2024', '23/02/2024', '12/03/2024', '13/03/2024'], ... 'C': ['1.5', 'N/A', '12.2', 'N/A'], ... }) >>> df A B C 0 one 02/02/2024 1.5 1 two 23/02/2024 N/A 2 two 12/03/2024 12.2 3 three 13/03/2024 N/A >>> df.dtypes A object B object C object dtype: object
>>> vectorizer = TableVectorizer() >>> vectorizer.fit_transform(df) A_one A_three A_two B_year B_month B_day B_total_seconds C 0 1.0 0.0 0.0 2024.0 2.0 2.0 1.706832e+09 1.5 1 0.0 0.0 1.0 2024.0 2.0 23.0 1.708646e+09 NaN 2 0.0 0.0 1.0 2024.0 3.0 12.0 1.710202e+09 12.2 3 0.0 1.0 0.0 2024.0 3.0 13.0 1.710288e+09 NaN
We can inspect which outputs were created from a given column in the input dataframe:
>>> vectorizer.input_to_outputs_['B'] ['B_year', 'B_month', 'B_day', 'B_total_seconds']
and the reverse mapping:
>>> vectorizer.output_to_input_['B_total_seconds'] 'B'
We can also see the encoder that was applied to a given column:
>>> vectorizer.transformers_['B'] DatetimeEncoder() >>> vectorizer.transformers_['A'] OneHotEncoder(drop='if_binary', dtype='float32', handle_unknown='ignore', sparse_output=False) >>> vectorizer.transformers_['A'].categories_ [array(['one', 'three', 'two'], dtype=object)]
We can see the columns grouped by the kind of encoder that was applied to them:
>>> vectorizer.kind_to_columns_ {'numeric': ['C'], 'datetime': ['B'], 'low_cardinality': ['A'], 'high_cardinality': [], 'specific': []}
As well as the reverse mapping (from each column to its kind):
>>> vectorizer.column_to_kind_ {'C': 'numeric', 'B': 'datetime', 'A': 'low_cardinality'}
Before applying the main transformer, the
TableVectorizerapplies several preprocessing steps, for example to detect numbers or dates that are represented as strings. Moreover, a final post-processing step is applied to all non-categorical columns in the encoder’s output to cast them to float32. We can inspect all the processing steps that were applied to a given column:>>> vectorizer.all_processing_steps_['B'] [CleanNullStrings(), ToDatetime(), DatetimeEncoder(), {'B_day': ToFloat32(), 'B_month': ToFloat32(), ...}]
Note that as the encoder (
DatetimeEncoder()above) produces multiple columns, the last processing step is not described by a single transformer like the previous ones but by a mapping from column name to transformer.all_processing_steps_is useful to inspect the details of the choices made by theTableVectorizerduring preprocessing, for example:>>> vectorizer.all_processing_steps_['B'][1] ToDatetime() >>> _.format_ '%d/%m/%Y'
Transformers are applied separately to each column
The
TableVectorizervectorizes each column separately – a different transformer is applied to each column; multivariate transformers are not allowed.>>> df_1 = pd.DataFrame(dict(A=['one', 'two'], B=['three', 'four'])) >>> vectorizer = TableVectorizer().fit(df_1) >>> vectorizer.transformers_['A'] is not vectorizer.transformers_['B'] True >>> vectorizer.transformers_['A'].categories_ [array(['one', 'two'], dtype=object)] >>> vectorizer.transformers_['B'].categories_ [array(['four', 'three'], dtype=object)]
Overriding the transformer for specific columns
We can also provide transformers for specific columns. In that case the provided transformer has full control over the associated columns; no other processing is applied to those columns. A column cannot appear twice in the
specific_transformers.Note
This functionality is likely to be removed in a future version of the
TableVectorizer.The overrides are provided as a list of pairs:
(transformer, list_of_column_names).>>> from sklearn.preprocessing import OrdinalEncoder >>> vectorizer = TableVectorizer( ... specific_transformers=[('drop', ['A']), (OrdinalEncoder(), ['B'])] ... ) >>> df A B C 0 one 02/02/2024 1.5 1 two 23/02/2024 N/A 2 two 12/03/2024 12.2 3 three 13/03/2024 N/A >>> vectorizer.fit_transform(df) B C 0 0.0 1.5 1 3.0 NaN 2 1.0 12.2 3 2.0 NaN
Here the column ‘A’ has been dropped and the column ‘B’ has been passed to the
OrdinalEncoder(instead of the default choice which would have beenDatetimeEncoder).We can see that ‘A’ and ‘B’ are now treated as ‘specific’ columns:
>>> vectorizer.column_to_kind_ {'C': 'numeric', 'A': 'specific', 'B': 'specific'}
Preprocessing and postprocessing steps are not applied to columns appearing in
specific_columns. For example ‘B’ has not gone throughToDatetime():>>> vectorizer.all_processing_steps_ {'A': [Drop()], 'B': [OrdinalEncoder()], 'C': [CleanNullStrings(), ToFloat32(), PassThrough(), {'C': ToFloat32()}]}
Specifying several
specific_transformersfor the same column is not allowed.>>> vectorizer = TableVectorizer( ... specific_transformers=[('passthrough', ['A', 'B']), ('drop', ['A'])] ... )
>>> vectorizer.fit_transform(df) Traceback (most recent call last): ... ValueError: Column 'A' used twice in 'specific_transformers', at indices 0 and 1.
- Attributes:
- transformers_
dict Maps the name of each column to the fitted transformer that was applied to it.
- column_to_kind_
dict Maps each column name to the kind (
"high_cardinality","low_cardinality","specific", etc.) it was assigned.- kind_to_columns_
dict The reverse of
column_to_kind_: maps each kind of column ("high_cardinality","low_cardinality", etc.) to a list of column names. For examplekind_to_columns['datetime']contains the names of all datetime columns.- input_to_outputs_
dict Maps the name of each input column to the names of the corresponding output columns.
- output_to_input_
dict The reverse of
input_to_outputs_: maps the name of each output column to the name of the column in the input dataframe from which it was derived.- all_processing_steps_
dict Maps the name of each column to a list of all the processing steps that were applied to it. Those steps may include some pre-processing transformations such as converting strings to datetimes or numbers, the main transformer (e.g. the
DatetimeEncoder), and a post-processing step casting the main transformer’s output to float32. See the “Examples” section below for details.- feature_names_in_
listof strings The names of the input columns, after applying some cleaning (casting all column names to strings and deduplication).
- n_features_in_
int The number of input columns.
- all_outputs_
listof strings The names of the output columns.
- transformers_
Methods
fit(X[, y])Fit transformer.
fit_transform(X[, y])Fit transformer and transform dataframe.
Return the column names of the output of
transformas a list of strings.Get metadata routing of this object.
get_params([deep])Get parameters for this estimator.
set_output(*[, transform])Set output container.
set_params(**params)Set the parameters of this estimator.
transform(X)Transform dataframe.
- fit(X, y=None)[source]#
Fit transformer.
- Parameters:
- Xdataframe of shape (n_samples, n_features)
Input data to transform.
- yarray_like, shape (n_samples,) or (n_samples, n_outputs) or
None, default=None Target values for supervised learning (None for unsupervised transformations).
- Returns:
- self
TableVectorizer The fitted estimator.
- self
- fit_transform(X, y=None)[source]#
Fit transformer and transform dataframe.
- Parameters:
- Xdataframe of shape (n_samples, n_features)
Input data to transform.
- yarray_like of shape (n_samples,) or (n_samples, n_outputs) or
None, default=None Target values for supervised learning (None for unsupervised transformations).
- Returns:
- dataframe
The transformed input.
- get_feature_names_out()[source]#
Return the column names of the output of
transformas a list of strings.- Returns:
listof stringsThe column names.
- get_metadata_routing()[source]#
Get metadata routing of this object.
Please check User Guide on how the routing mechanism works.
- Returns:
- routingMetadataRequest
A
MetadataRequestencapsulating routing information.
- set_output(*, transform=None)[source]#
Set output container.
See Introducing the set_output API for an example on how to use the API.
- Parameters:
- transform{“default”, “pandas”, “polars”}, default=None
Configure output of transform and fit_transform.
“default”: Default output format of a transformer
“pandas”: DataFrame output
“polars”: Polars output
None: Transform configuration is unchanged
Added in version 1.4: “polars” option was added.
- Returns:
- selfestimator instance
Estimator instance.
- set_params(**params)[source]#
Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as
Pipeline). The latter have parameters of the form<component>__<parameter>so that it’s possible to update each component of a nested object.- Parameters:
- **params
dict Estimator parameters.
- **params
- Returns:
- selfestimator instance
Estimator instance.
Examples using skrub.TableVectorizer#
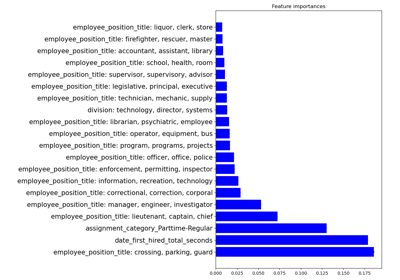
Encoding: from a dataframe to a numerical matrix for machine learning
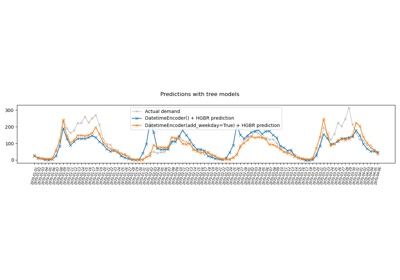
Handling datetime features with the DatetimeEncoder
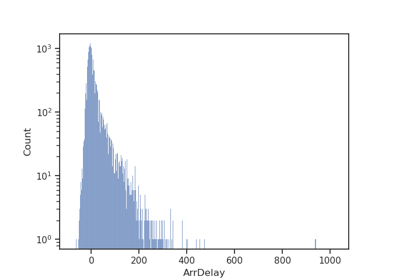
Spatial join for flight data: Joining across multiple columns
