Note
Go to the end to download the full example code. or to run this example in your browser via JupyterLite or Binder
Various string encoders: a sentiment analysis example#
In this example, we explore the performance of string and categorical encoders available in skrub.
The Toxicity dataset#
We focus on the toxicity dataset, a corpus of 1,000 tweets, evenly balanced between the binary labels “Toxic” and “Not Toxic”. Our goal is to classify each entry between these two labels, using only the text of the tweets as features.
from skrub.datasets import fetch_toxicity
dataset = fetch_toxicity()
X, y = dataset.X, dataset.y
X["is_toxic"] = y
Downloading 'toxicity' from https://github.com/skrub-data/skrub-data-files/raw/refs/heads/main/toxicity.zip (attempt 1/3)
When it comes to displaying large chunks of text, the TableReport is especially
useful! Click on any cell below to expand and read the tweet in full.
from skrub import TableReport
TableReport(X)
| text | is_toxic | |
|---|---|---|
| 0 | Elon Musk is a piece of shit, greedy capitalist who exploits workers, and offers nothing of real benefit to the world. All he’s done is make a name for himself on the backs of other people, using dirty money from his family’s emerald mine they acquired during apartheid. I don’t care that he’s autistic. He thinks we should be cured with his company’s AI chip. He is not a representation of our community. Don’t celebrate him on this page. | Toxic |
| 1 | The senile credit card shrill from Delaware needs to resign!! | Toxic |
| 2 | He does that a lot -- makes everyone look good but him...I guess it's also probably the Dems and the Media that force him to compulsively tweet abject bullshit like a lying bitch. They're tricky, them libs. | Toxic |
| 3 | F*ck Lizzo | Toxic |
| 4 | Epstein and trump were best buds!!! Pedophiles who play together!! | Toxic |
| 995 | My maternal abuelita taught me how to make plantain empanadas 🥺 and my paternal abuelita needed me to help her brush her dentures 😌 I love them so much 🥰 | Not Toxic |
| 996 | Funnily enough I was looking online last week and wondering why nobody has opened an eSports/Gaming bar round here. Can’t wait to pop in at some point :) | Not Toxic |
| 997 | I can't bear how nice this is. I guess its bearnessities. I'll see my self out | Not Toxic |
| 998 | Going to buy a share of Tesla just to ensure it starts going back down | Not Toxic |
| 999 | I only saw a couple of these throughout the month and tried to figure out what all of them were. Only ones I missed were Star Guardian Seraphine (thought it was Heartbreaker) and I couldn't figure out the 2nd Soraka was Victorious. So all in all, you did a really good job nailing the characters and the theme presented! I think my faves are KDA Neeko and CCN Xayah. | Not Toxic |
text
ObjectDType- Null values
- 0 (0.0%)
- Unique values
-
999 (99.9%)
This column has a high cardinality (> 40).
Most frequent values
Post was funny, but this took it to another level.
I think Seju drops 2 as well (from Bristle) Not sure about Quinn tho...
Great depth! This is wicked
Love it! Thank you for finding me a new sub to join.
I was playing a rogue when I came up with the idea. exactly it's intended use!
This happened to me too when I did razor's quest. But only his though, after that it went back to normal lmao
Interesting survey, already answered :)
The Furies of Calderon series is on my to-read list. I love his Harry Dresden series so I'm hopeful.
Not too late to contact Sky News and heap your own media pressure on the Premier.
List:There is no chance that MBTL ends up as difficult to find matches as Current Code in the next decade. The game has rollback netcode. Don't make the mistake of focusing on how many people are online
['Post was funny, but this took it to another level.', 'I think Seju drops 2 as well (from Bristle) Not sure about Quinn tho...', 'Great depth! This is wicked', 'Love it! Thank you for finding me a new sub to join.', "I was playing a rogue when I came up with the idea. exactly it's intended use!", "This happened to me too when I did razor's quest. But only his though, after that it went back to normal lmao", 'Interesting survey, already answered :)', "The Furies of Calderon series is on my to-read list. I love his Harry Dresden series so I'm hopeful.", 'Not too late to contact Sky News and heap your own media pressure on the Premier.', "There is no chance that MBTL ends up as difficult to find matches as Current Code in the next decade. The game has rollback netcode. Don't make the mistake of focusing on how many people are online"]
is_toxic
ObjectDType- Null values
- 0 (0.0%)
- Unique values
- 2 (0.2%)
Most frequent values
Toxic
Not Toxic
['Toxic', 'Not Toxic']
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
|
Column
|
Column name
|
dtype
|
Is sorted
|
Null values
|
Unique values
|
Mean
|
Std
|
Min
|
Median
|
Max
|
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | text | ObjectDType | False | 0 (0.0%) | 999 (99.9%) | |||||
| 1 | is_toxic | ObjectDType | True | 0 (0.0%) | 2 (0.2%) |
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
text
ObjectDType- Null values
- 0 (0.0%)
- Unique values
-
999 (99.9%)
This column has a high cardinality (> 40).
Most frequent values
Post was funny, but this took it to another level.
I think Seju drops 2 as well (from Bristle) Not sure about Quinn tho...
Great depth! This is wicked
Love it! Thank you for finding me a new sub to join.
I was playing a rogue when I came up with the idea. exactly it's intended use!
This happened to me too when I did razor's quest. But only his though, after that it went back to normal lmao
Interesting survey, already answered :)
The Furies of Calderon series is on my to-read list. I love his Harry Dresden series so I'm hopeful.
Not too late to contact Sky News and heap your own media pressure on the Premier.
List:There is no chance that MBTL ends up as difficult to find matches as Current Code in the next decade. The game has rollback netcode. Don't make the mistake of focusing on how many people are online
['Post was funny, but this took it to another level.', 'I think Seju drops 2 as well (from Bristle) Not sure about Quinn tho...', 'Great depth! This is wicked', 'Love it! Thank you for finding me a new sub to join.', "I was playing a rogue when I came up with the idea. exactly it's intended use!", "This happened to me too when I did razor's quest. But only his though, after that it went back to normal lmao", 'Interesting survey, already answered :)', "The Furies of Calderon series is on my to-read list. I love his Harry Dresden series so I'm hopeful.", 'Not too late to contact Sky News and heap your own media pressure on the Premier.', "There is no chance that MBTL ends up as difficult to find matches as Current Code in the next decade. The game has rollback netcode. Don't make the mistake of focusing on how many people are online"]
is_toxic
ObjectDType- Null values
- 0 (0.0%)
- Unique values
- 2 (0.2%)
Most frequent values
Toxic
Not Toxic
['Toxic', 'Not Toxic']
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
| Column 1 | Column 2 | Cramér's V | Pearson's Correlation |
|---|---|---|---|
| text | is_toxic | 0.100 |
Please enable javascript
The skrub table reports need javascript to display correctly. If you are displaying a report in a Jupyter notebook and you see this message, you may need to re-execute the cell or to trust the notebook (button on the top right or "File > Trust notebook").
GapEncoder#
First, let’s vectorize our text column using the GapEncoder, one of the
high cardinality categorical encoders
provided by skrub.
As introduced in the previous example, the GapEncoder
performs matrix factorization for topic modeling. It builds latent topics by
capturing combinations of substrings that frequently co-occur, and encoded vectors
correspond to topic activations.
To interpret these latent topics, we select for each of them a few labels from the input data with the highest activations. In the example below we select 3 labels to summarize each topic.
from skrub import GapEncoder
gap = GapEncoder(n_components=30)
X_trans = gap.fit_transform(X["text"])
# Add the original text as a first column
X_trans.insert(0, "text", X["text"])
TableReport(X_trans)
| text | text: nvrseqrvrqrqrr, highlights, might | text: suicide, fucking, joking | text: atlantic, across, cross | text: participation, redundant, repeatedly | text: politicians, politician, amazing | text: pedophile, pedophiles, pfffffft | text: congress, blackmailed, prosecuting | text: democraps, leadership, democrap | text: afghanistan, withdrawal, campaigned | text: purchasing, purchased, transaction | text: goinggoinggoing, awkwardly, occasionally | text: lackluster, survivability, shyvana | text: interview, interviews, japanese | text: pizzaaaa, ƞỉဌဌᕦѓ, utm_medium | text: approachable, foreigners, friendly | text: yourselves, ourselves, unapologetic | text: government, governments, destroying | text: trumptards, exercises, exercise | text: definitely, considerable, considered | text: deepening, between, widening | text: liberalhypocrisy, boycotthollywood, hypocrisy | text: announcement, experienced, experience | text: people, fauci, peoples | text: legitimate, capitalists, capitalist | text: gargantuan, emblazoned, confederate | text: ridiculousness, ridiculous, subtract | text: corporate, doomberg, because | text: celebrities, culture, happened | text: qualified, marxists, americans | text: vaccinated, airlines, vaccines | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Elon Musk is a piece of shit, greedy capitalist who exploits workers, and offers nothing of real benefit to the world. All he’s done is make a name for himself on the backs of other people, using dirty money from his family’s emerald mine they acquired during apartheid. I don’t care that he’s autistic. He thinks we should be cured with his company’s AI chip. He is not a representation of our community. Don’t celebrate him on this page. | 0.00205 | 0.0122 | 0.000890 | 0.0839 | 0.00353 | 0.000686 | 0.124 | 0.000883 | 0.108 | 0.0645 | 0.00768 | 0.0146 | 0.00386 | 0.000719 | 0.00358 | 0.00253 | 0.521 | 0.0182 | 0.350 | 0.00103 | 0.0114 | 0.0478 | 0.210 | 0.681 | 0.0308 | 0.790 | 0.00151 | 0.150 | 0.0446 | 0.00154 |
| 1 | The senile credit card shrill from Delaware needs to resign!! | 0.000694 | 0.00412 | 0.000752 | 0.00165 | 0.000482 | 0.00340 | 0.0170 | 0.000878 | 0.00102 | 0.0394 | 0.00785 | 0.00462 | 0.00501 | 0.000639 | 0.0612 | 0.00148 | 0.000989 | 0.00231 | 0.00117 | 0.00401 | 0.00338 | 0.0436 | 0.0105 | 0.0194 | 0.00298 | 0.208 | 0.00123 | 0.00136 | 0.00151 | 0.000829 |
| 2 | He does that a lot -- makes everyone look good but him...I guess it's also probably the Dems and the Media that force him to compulsively tweet abject bullshit like a lying bitch. They're tricky, them libs. | 0.00220 | 0.498 | 0.165 | 0.00129 | 0.00269 | 0.000894 | 0.00611 | 0.0642 | 0.000870 | 0.00101 | 0.000766 | 0.0993 | 0.00161 | 0.000978 | 0.000927 | 0.00207 | 0.0194 | 0.0712 | 0.0823 | 0.178 | 0.00356 | 0.0317 | 0.0210 | 0.0131 | 0.0801 | 0.0144 | 0.0142 | 0.0127 | 0.00363 | 0.148 |
| 3 | F*ck Lizzo | 0.0493 | 0.00369 | 0.000383 | 0.000362 | 0.000355 | 0.000514 | 0.000348 | 0.000358 | 0.000413 | 0.000306 | 0.000301 | 0.000337 | 0.000337 | 0.00265 | 0.000309 | 0.000434 | 0.000410 | 0.000323 | 0.000280 | 0.000423 | 0.000790 | 0.000354 | 0.000438 | 0.000319 | 0.000374 | 0.000282 | 0.00106 | 0.00104 | 0.000326 | 0.000786 |
| 4 | Epstein and trump were best buds!!! Pedophiles who play together!! | 0.000375 | 0.000835 | 0.000507 | 0.00136 | 0.000451 | 0.238 | 0.0320 | 0.000655 | 0.0144 | 0.000565 | 0.00102 | 0.00222 | 0.000707 | 0.000485 | 0.0227 | 0.0923 | 0.00491 | 0.00722 | 0.00214 | 0.000871 | 0.00443 | 0.00127 | 0.000899 | 0.000608 | 0.0226 | 0.00133 | 0.000584 | 0.0123 | 0.0202 | 0.000993 |
| 995 | My maternal abuelita taught me how to make plantain empanadas 🥺 and my paternal abuelita needed me to help her brush her dentures 😌 I love them so much 🥰 | 0.00101 | 0.00180 | 0.00409 | 0.0508 | 0.443 | 0.000606 | 0.00135 | 0.00623 | 0.109 | 0.00696 | 0.00107 | 0.151 | 0.00143 | 0.000732 | 0.0824 | 0.00133 | 0.0384 | 0.000763 | 0.00608 | 0.0181 | 0.00220 | 0.00242 | 0.000810 | 0.0190 | 0.123 | 0.0399 | 0.000941 | 0.0234 | 0.00365 | 0.000829 |
| 996 | Funnily enough I was looking online last week and wondering why nobody has opened an eSports/Gaming bar round here. Can’t wait to pop in at some point :) | 0.00160 | 0.102 | 0.000672 | 0.0520 | 0.113 | 0.000569 | 0.0669 | 0.0298 | 0.0646 | 0.0252 | 0.208 | 0.00155 | 0.303 | 0.000636 | 0.000766 | 0.00198 | 0.00456 | 0.0333 | 0.0303 | 0.00509 | 0.00126 | 0.00329 | 0.000896 | 0.000571 | 0.00506 | 0.000717 | 0.00130 | 0.0673 | 0.00354 | 0.0125 |
| 997 | I can't bear how nice this is. I guess its bearnessities. I'll see my self out | 0.00102 | 0.000675 | 0.00192 | 0.00117 | 0.0368 | 0.000607 | 0.0276 | 0.00483 | 0.00626 | 0.0607 | 0.000565 | 0.145 | 0.0730 | 0.00103 | 0.00129 | 0.119 | 0.00108 | 0.00221 | 0.00103 | 0.0289 | 0.00239 | 0.00278 | 0.00480 | 0.00113 | 0.000678 | 0.0212 | 0.000830 | 0.0271 | 0.00251 | 0.00109 |
| 998 | Going to buy a share of Tesla just to ensure it starts going back down | 0.00539 | 0.00582 | 0.000498 | 0.0507 | 0.000899 | 0.000474 | 0.000854 | 0.00831 | 0.000703 | 0.000870 | 0.259 | 0.0836 | 0.000555 | 0.000474 | 0.00120 | 0.000830 | 0.00107 | 0.0154 | 0.00186 | 0.00503 | 0.000681 | 0.000687 | 0.000637 | 0.000715 | 0.0442 | 0.000873 | 0.00715 | 0.00231 | 0.0109 | 0.00648 |
| 999 | I only saw a couple of these throughout the month and tried to figure out what all of them were. Only ones I missed were Star Guardian Seraphine (thought it was Heartbreaker) and I couldn't figure out the 2nd Soraka was Victorious. So all in all, you did a really good job nailing the characters and the theme presented! I think my faves are KDA Neeko and CCN Xayah. | 0.0167 | 0.0288 | 0.00210 | 0.00246 | 0.0417 | 0.0245 | 0.288 | 0.00146 | 0.277 | 0.174 | 0.00694 | 0.100 | 0.441 | 0.00158 | 0.00126 | 0.0681 | 0.00326 | 0.331 | 0.0616 | 0.00782 | 0.171 | 0.00114 | 0.0282 | 0.0412 | 0.136 | 0.0199 | 0.00359 | 0.00583 | 0.176 | 0.283 |
text
ObjectDType- Null values
- 0 (0.0%)
- Unique values
-
999 (99.9%)
This column has a high cardinality (> 40).
text: nvrseqrvrqrqrr, highlights, might
Float64DType- Null values
- 0 (0.0%)
- Unique values
-
999 (99.9%)
This column has a high cardinality (> 40).
- Mean ± Std
- 0.0185 ± 0.0606
- Median ± IQR
- 0.000864 ± 0.00188
- Min | Max
- 0.000256 | 0.648
text: suicide, fucking, joking
Float64DType- Null values
- 0 (0.0%)
- Unique values
-
999 (99.9%)
This column has a high cardinality (> 40).
- Mean ± Std
- 0.0476 ± 0.207
- Median ± IQR
- 0.00569 ± 0.0509
- Min | Max
- 0.000253 | 6.13
text: atlantic, across, cross
Float64DType- Null values
- 0 (0.0%)
- Unique values
-
999 (99.9%)
This column has a high cardinality (> 40).
- Mean ± Std
- 0.0158 ± 0.0749
- Median ± IQR
- 0.000829 ± 0.00114
- Min | Max
- 0.000264 | 1.25
text: participation, redundant, repeatedly
Float64DType- Null values
- 0 (0.0%)
- Unique values
-
999 (99.9%)
This column has a high cardinality (> 40).
- Mean ± Std
- 0.0317 ± 0.140
- Median ± IQR
- 0.00241 ± 0.0221
- Min | Max
- 0.000253 | 3.96
text: politicians, politician, amazing
Float64DType- Null values
- 0 (0.0%)
- Unique values
-
999 (99.9%)
This column has a high cardinality (> 40).
- Mean ± Std
- 0.0227 ± 0.0737
- Median ± IQR
- 0.00107 ± 0.00387
- Min | Max
- 0.000269 | 0.820
text: pedophile, pedophiles, pfffffft
Float64DType- Null values
- 0 (0.0%)
- Unique values
-
999 (99.9%)
This column has a high cardinality (> 40).
- Mean ± Std
- 0.00977 ± 0.0942
- Median ± IQR
- 0.000645 ± 0.000527
- Min | Max
- 0.000260 | 2.48
text: congress, blackmailed, prosecuting
Float64DType- Null values
- 0 (0.0%)
- Unique values
-
999 (99.9%)
This column has a high cardinality (> 40).
- Mean ± Std
- 0.0499 ± 0.352
- Median ± IQR
- 0.00397 ± 0.0346
- Min | Max
- 0.000253 | 10.8
text: democraps, leadership, democrap
Float64DType- Null values
- 0 (0.0%)
- Unique values
-
999 (99.9%)
This column has a high cardinality (> 40).
- Mean ± Std
- 0.0260 ± 0.0782
- Median ± IQR
- 0.000968 ± 0.00755
- Min | Max
- 0.000272 | 0.920
text: afghanistan, withdrawal, campaigned
Float64DType- Null values
- 0 (0.0%)
- Unique values
-
999 (99.9%)
This column has a high cardinality (> 40).
- Mean ± Std
- 0.0377 ± 0.198
- Median ± IQR
- 0.00258 ± 0.0296
- Min | Max
- 0.000256 | 5.89
text: purchasing, purchased, transaction
Float64DType- Null values
- 0 (0.0%)
- Unique values
-
999 (99.9%)
This column has a high cardinality (> 40).
- Mean ± Std
- 0.0258 ± 0.0924
- Median ± IQR
- 0.00134 ± 0.0105
- Min | Max
- 0.000255 | 1.72
text: goinggoinggoing, awkwardly, occasionally
Float64DType- Null values
- 0 (0.0%)
- Unique values
-
999 (99.9%)
This column has a high cardinality (> 40).
- Mean ± Std
- 0.0303 ± 0.0904
- Median ± IQR
- 0.00145 ± 0.0140
- Min | Max
- 0.000256 | 1.55
text: lackluster, survivability, shyvana
Float64DType- Null values
- 0 (0.0%)
- Unique values
-
999 (99.9%)
This column has a high cardinality (> 40).
- Mean ± Std
- 0.0545 ± 0.454
- Median ± IQR
- 0.00358 ± 0.0323
- Min | Max
- 0.000252 | 14.1
text: interview, interviews, japanese
Float64DType- Null values
- 0 (0.0%)
- Unique values
-
999 (99.9%)
This column has a high cardinality (> 40).
- Mean ± Std
- 0.0412 ± 0.148
- Median ± IQR
- 0.00177 ± 0.0271
- Min | Max
- 0.000255 | 3.07
text: pizzaaaa, ƞỉဌဌᕦѓ, utm_medium
Float64DType- Null values
- 0 (0.0%)
- Unique values
-
999 (99.9%)
This column has a high cardinality (> 40).
- Mean ± Std
- 0.0180 ± 0.111
- Median ± IQR
- 0.000638 ± 0.000509
- Min | Max
- 0.000258 | 1.64
text: approachable, foreigners, friendly
Float64DType- Null values
- 0 (0.0%)
- Unique values
-
999 (99.9%)
This column has a high cardinality (> 40).
- Mean ± Std
- 0.0236 ± 0.0777
- Median ± IQR
- 0.00103 ± 0.00679
- Min | Max
- 0.000257 | 1.23
text: yourselves, ourselves, unapologetic
Float64DType- Null values
- 0 (0.0%)
- Unique values
-
999 (99.9%)
This column has a high cardinality (> 40).
- Mean ± Std
- 0.0593 ± 0.454
- Median ± IQR
- 0.00273 ± 0.0382
- Min | Max
- 0.000252 | 14.0
text: government, governments, destroying
Float64DType- Null values
- 0 (0.0%)
- Unique values
-
999 (99.9%)
This column has a high cardinality (> 40).
- Mean ± Std
- 0.0537 ± 0.422
- Median ± IQR
- 0.00318 ± 0.0298
- Min | Max
- 0.000251 | 13.0
text: trumptards, exercises, exercise
Float64DType- Null values
- 0 (0.0%)
- Unique values
-
999 (99.9%)
This column has a high cardinality (> 40).
- Mean ± Std
- 0.0334 ± 0.120
- Median ± IQR
- 0.00166 ± 0.0160
- Min | Max
- 0.000254 | 2.87
text: definitely, considerable, considered
Float64DType- Null values
- 0 (0.0%)
- Unique values
-
999 (99.9%)
This column has a high cardinality (> 40).
- Mean ± Std
- 0.0472 ± 0.217
- Median ± IQR
- 0.00282 ± 0.0353
- Min | Max
- 0.000254 | 6.17
text: deepening, between, widening
Float64DType- Null values
- 0 (0.0%)
- Unique values
-
999 (99.9%)
This column has a high cardinality (> 40).
- Mean ± Std
- 0.0329 ± 0.141
- Median ± IQR
- 0.00293 ± 0.0262
- Min | Max
- 0.000254 | 4.01
text: liberalhypocrisy, boycotthollywood, hypocrisy
Float64DType- Null values
- 0 (0.0%)
- Unique values
-
999 (99.9%)
This column has a high cardinality (> 40).
- Mean ± Std
- 0.0292 ± 0.111
- Median ± IQR
- 0.00158 ± 0.0137
- Min | Max
- 0.000258 | 2.68
text: announcement, experienced, experience
Float64DType- Null values
- 0 (0.0%)
- Unique values
-
999 (99.9%)
This column has a high cardinality (> 40).
- Mean ± Std
- 0.0312 ± 0.117
- Median ± IQR
- 0.00180 ± 0.0179
- Min | Max
- 0.000254 | 2.89
text: people, fauci, peoples
Float64DType- Null values
- 0 (0.0%)
- Unique values
-
999 (99.9%)
This column has a high cardinality (> 40).
- Mean ± Std
- 0.0203 ± 0.0646
- Median ± IQR
- 0.000874 ± 0.00258
- Min | Max
- 0.000258 | 0.650
text: legitimate, capitalists, capitalist
Float64DType- Null values
- 0 (0.0%)
- Unique values
-
999 (99.9%)
This column has a high cardinality (> 40).
- Mean ± Std
- 0.0342 ± 0.139
- Median ± IQR
- 0.00189 ± 0.0214
- Min | Max
- 0.000256 | 3.59
text: gargantuan, emblazoned, confederate
Float64DType- Null values
- 0 (0.0%)
- Unique values
-
999 (99.9%)
This column has a high cardinality (> 40).
- Mean ± Std
- 0.0481 ± 0.368
- Median ± IQR
- 0.00381 ± 0.0369
- Min | Max
- 0.000253 | 11.4
text: ridiculousness, ridiculous, subtract
Float64DType- Null values
- 0 (0.0%)
- Unique values
-
999 (99.9%)
This column has a high cardinality (> 40).
- Mean ± Std
- 0.0263 ± 0.0877
- Median ± IQR
- 0.00160 ± 0.0127
- Min | Max
- 0.000255 | 1.77
text: corporate, doomberg, because
Float64DType- Null values
- 0 (0.0%)
- Unique values
-
999 (99.9%)
This column has a high cardinality (> 40).
- Mean ± Std
- 0.0335 ± 0.150
- Median ± IQR
- 0.00191 ± 0.0231
- Min | Max
- 0.000254 | 4.27
text: celebrities, culture, happened
Float64DType- Null values
- 0 (0.0%)
- Unique values
-
999 (99.9%)
This column has a high cardinality (> 40).
- Mean ± Std
- 0.0305 ± 0.0987
- Median ± IQR
- 0.00197 ± 0.0259
- Min | Max
- 0.000256 | 2.26
text: qualified, marxists, americans
Float64DType- Null values
- 0 (0.0%)
- Unique values
-
999 (99.9%)
This column has a high cardinality (> 40).
- Mean ± Std
- 0.0445 ± 0.247
- Median ± IQR
- 0.00363 ± 0.0315
- Min | Max
- 0.000253 | 7.42
text: vaccinated, airlines, vaccines
Float64DType- Null values
- 0 (0.0%)
- Unique values
-
999 (99.9%)
This column has a high cardinality (> 40).
- Mean ± Std
- 0.0295 ± 0.162
- Median ± IQR
- 0.00231 ± 0.0171
- Min | Max
- 0.000255 | 4.81
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
|
Column
|
Column name
|
dtype
|
Is sorted
|
Null values
|
Unique values
|
Mean
|
Std
|
Min
|
Median
|
Max
|
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | text | ObjectDType | False | 0 (0.0%) | 999 (99.9%) | |||||
| 1 | text: nvrseqrvrqrqrr, highlights, might | Float64DType | False | 0 (0.0%) | 999 (99.9%) | 0.0185 | 0.0606 | 0.000256 | 0.000864 | 0.648 |
| 2 | text: suicide, fucking, joking | Float64DType | False | 0 (0.0%) | 999 (99.9%) | 0.0476 | 0.207 | 0.000253 | 0.00569 | 6.13 |
| 3 | text: atlantic, across, cross | Float64DType | False | 0 (0.0%) | 999 (99.9%) | 0.0158 | 0.0749 | 0.000264 | 0.000829 | 1.25 |
| 4 | text: participation, redundant, repeatedly | Float64DType | False | 0 (0.0%) | 999 (99.9%) | 0.0317 | 0.140 | 0.000253 | 0.00241 | 3.96 |
| 5 | text: politicians, politician, amazing | Float64DType | False | 0 (0.0%) | 999 (99.9%) | 0.0227 | 0.0737 | 0.000269 | 0.00107 | 0.820 |
| 6 | text: pedophile, pedophiles, pfffffft | Float64DType | False | 0 (0.0%) | 999 (99.9%) | 0.00977 | 0.0942 | 0.000260 | 0.000645 | 2.48 |
| 7 | text: congress, blackmailed, prosecuting | Float64DType | False | 0 (0.0%) | 999 (99.9%) | 0.0499 | 0.352 | 0.000253 | 0.00397 | 10.8 |
| 8 | text: democraps, leadership, democrap | Float64DType | False | 0 (0.0%) | 999 (99.9%) | 0.0260 | 0.0782 | 0.000272 | 0.000968 | 0.920 |
| 9 | text: afghanistan, withdrawal, campaigned | Float64DType | False | 0 (0.0%) | 999 (99.9%) | 0.0377 | 0.198 | 0.000256 | 0.00258 | 5.89 |
| 10 | text: purchasing, purchased, transaction | Float64DType | False | 0 (0.0%) | 999 (99.9%) | 0.0258 | 0.0924 | 0.000255 | 0.00134 | 1.72 |
| 11 | text: goinggoinggoing, awkwardly, occasionally | Float64DType | False | 0 (0.0%) | 999 (99.9%) | 0.0303 | 0.0904 | 0.000256 | 0.00145 | 1.55 |
| 12 | text: lackluster, survivability, shyvana | Float64DType | False | 0 (0.0%) | 999 (99.9%) | 0.0545 | 0.454 | 0.000252 | 0.00358 | 14.1 |
| 13 | text: interview, interviews, japanese | Float64DType | False | 0 (0.0%) | 999 (99.9%) | 0.0412 | 0.148 | 0.000255 | 0.00177 | 3.07 |
| 14 | text: pizzaaaa, ƞỉဌဌᕦѓ, utm_medium | Float64DType | False | 0 (0.0%) | 999 (99.9%) | 0.0180 | 0.111 | 0.000258 | 0.000638 | 1.64 |
| 15 | text: approachable, foreigners, friendly | Float64DType | False | 0 (0.0%) | 999 (99.9%) | 0.0236 | 0.0777 | 0.000257 | 0.00103 | 1.23 |
| 16 | text: yourselves, ourselves, unapologetic | Float64DType | False | 0 (0.0%) | 999 (99.9%) | 0.0593 | 0.454 | 0.000252 | 0.00273 | 14.0 |
| 17 | text: government, governments, destroying | Float64DType | False | 0 (0.0%) | 999 (99.9%) | 0.0537 | 0.422 | 0.000251 | 0.00318 | 13.0 |
| 18 | text: trumptards, exercises, exercise | Float64DType | False | 0 (0.0%) | 999 (99.9%) | 0.0334 | 0.120 | 0.000254 | 0.00166 | 2.87 |
| 19 | text: definitely, considerable, considered | Float64DType | False | 0 (0.0%) | 999 (99.9%) | 0.0472 | 0.217 | 0.000254 | 0.00282 | 6.17 |
| 20 | text: deepening, between, widening | Float64DType | False | 0 (0.0%) | 999 (99.9%) | 0.0329 | 0.141 | 0.000254 | 0.00293 | 4.01 |
| 21 | text: liberalhypocrisy, boycotthollywood, hypocrisy | Float64DType | False | 0 (0.0%) | 999 (99.9%) | 0.0292 | 0.111 | 0.000258 | 0.00158 | 2.68 |
| 22 | text: announcement, experienced, experience | Float64DType | False | 0 (0.0%) | 999 (99.9%) | 0.0312 | 0.117 | 0.000254 | 0.00180 | 2.89 |
| 23 | text: people, fauci, peoples | Float64DType | False | 0 (0.0%) | 999 (99.9%) | 0.0203 | 0.0646 | 0.000258 | 0.000874 | 0.650 |
| 24 | text: legitimate, capitalists, capitalist | Float64DType | False | 0 (0.0%) | 999 (99.9%) | 0.0342 | 0.139 | 0.000256 | 0.00189 | 3.59 |
| 25 | text: gargantuan, emblazoned, confederate | Float64DType | False | 0 (0.0%) | 999 (99.9%) | 0.0481 | 0.368 | 0.000253 | 0.00381 | 11.4 |
| 26 | text: ridiculousness, ridiculous, subtract | Float64DType | False | 0 (0.0%) | 999 (99.9%) | 0.0263 | 0.0877 | 0.000255 | 0.00160 | 1.77 |
| 27 | text: corporate, doomberg, because | Float64DType | False | 0 (0.0%) | 999 (99.9%) | 0.0335 | 0.150 | 0.000254 | 0.00191 | 4.27 |
| 28 | text: celebrities, culture, happened | Float64DType | False | 0 (0.0%) | 999 (99.9%) | 0.0305 | 0.0987 | 0.000256 | 0.00197 | 2.26 |
| 29 | text: qualified, marxists, americans | Float64DType | False | 0 (0.0%) | 999 (99.9%) | 0.0445 | 0.247 | 0.000253 | 0.00363 | 7.42 |
| 30 | text: vaccinated, airlines, vaccines | Float64DType | False | 0 (0.0%) | 999 (99.9%) | 0.0295 | 0.162 | 0.000255 | 0.00231 | 4.81 |
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
max_plot_columns
limit set for the TableReport during report creation.
No columns match the selected filter: . You can change the column filter in the dropdown menu above.
max_association_columns parameter.
Please enable javascript
The skrub table reports need javascript to display correctly. If you are displaying a report in a Jupyter notebook and you see this message, you may need to re-execute the cell or to trust the notebook (button on the top right or "File > Trust notebook").
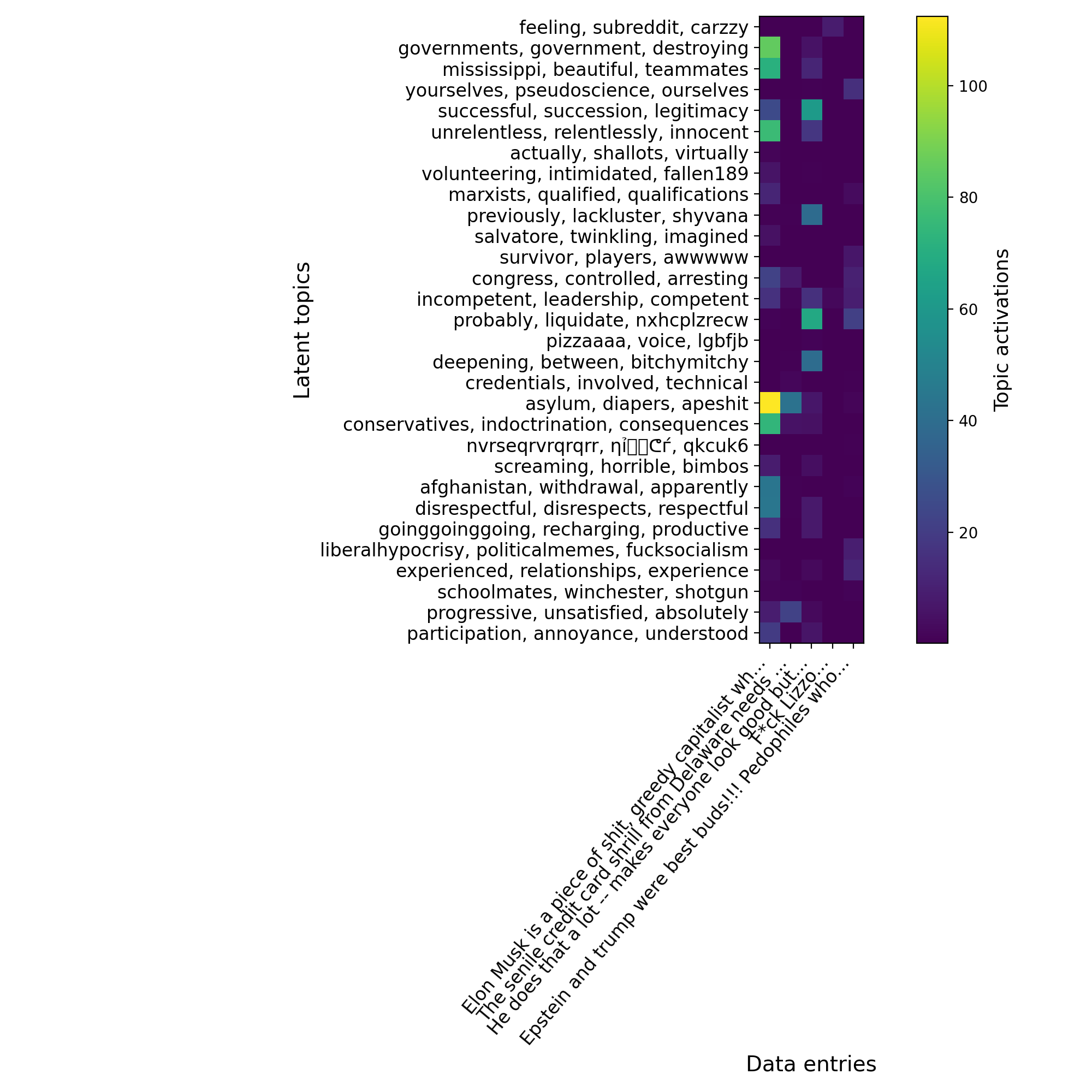
We can use a heatmap to highlight the highest activations, making them more visible for comparison against the original text and vectors above.
import numpy as np
from matplotlib import pyplot as plt
def plot_gap_feature_importance(X_trans):
x_samples = X_trans.pop("text")
# We slightly format the topics and labels for them to fit on the plot.
topic_labels = [x.replace("text: ", "") for x in X_trans.columns]
labels = x_samples.str[:50].values + "..."
# We clip large outliers to make activations more visible.
X_trans = np.clip(X_trans, a_min=None, a_max=200)
plt.figure(figsize=(10, 10), dpi=200)
plt.imshow(X_trans.T)
plt.yticks(
range(len(topic_labels)),
labels=topic_labels,
ha="right",
size=12,
)
plt.xticks(range(len(labels)), labels=labels, size=12, rotation=50, ha="right")
plt.colorbar().set_label(label="Topic activations", size=13)
plt.ylabel("Latent topics", size=14)
plt.xlabel("Data entries", size=14)
plt.tight_layout()
plt.show()
plot_gap_feature_importance(X_trans.head())
/home/circleci/project/examples/02_text_with_string_encoders.py:116: UserWarning:
Glyph 4108 (\N{MYANMAR LETTER TTHA}) missing from font(s) DejaVu Sans.
Now that we have an understanding of the vectors produced by the GapEncoder,
let’s evaluate its performance in toxicity classification. The GapEncoder excels
at handling categorical columns with high cardinality, but here the column consists
of free-form text. Sentences are generally longer, with more unique ngrams than
high cardinality categories.
To benchmark the performance of the GapEncoder against the toxicity dataset,
we integrate it into a TableVectorizer, as introduced in the
previous example,
and create a Pipeline by appending a HistGradientBoostingClassifier, which
consumes the vectors produced by the GapEncoder.
We set n_components to 30; however, to achieve the best performance, we would
need to find the optimal value for this hyperparameter using either GridSearchCV
or RandomizedSearchCV. We skip this part to keep the computation time for this
small example.
Recall that the ROC AUC is a metric that quantifies the ranking power of estimators, where a random estimator scores 0.5, and an oracle —providing perfect predictions— scores 1.
from sklearn.ensemble import HistGradientBoostingClassifier
from sklearn.model_selection import cross_validate
from sklearn.pipeline import make_pipeline
from skrub import TableVectorizer
def plot_box_results(named_results):
fig, ax = plt.subplots()
names, scores = zip(
*[(name, result["test_score"]) for name, result in named_results]
)
ax.boxplot(scores)
ax.set_xticks(range(1, len(names) + 1), labels=list(names), size=12)
ax.set_ylabel("ROC AUC", size=14)
plt.title(
"AUC distribution across folds (higher is better)",
size=14,
)
plt.show()
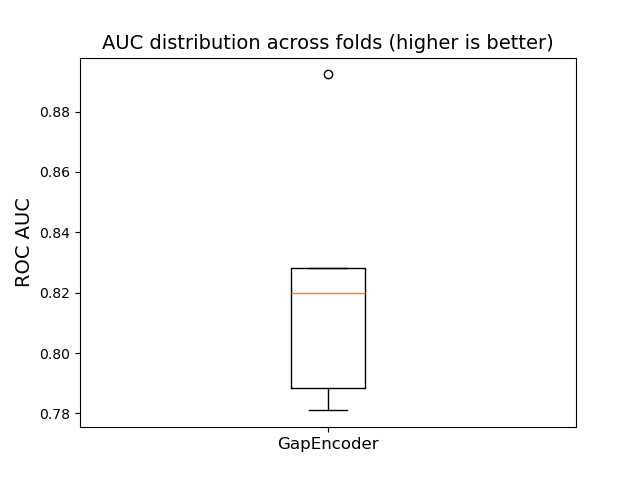
results = []
y = X.pop("is_toxic").map({"Toxic": 1, "Not Toxic": 0})
gap_pipe = make_pipeline(
TableVectorizer(high_cardinality=GapEncoder(n_components=30)),
HistGradientBoostingClassifier(),
)
gap_results = cross_validate(gap_pipe, X, y, scoring="roc_auc")
results.append(("GapEncoder", gap_results))
plot_box_results(results)
MinHashEncoder#
We now compare these results with the MinHashEncoder, which is faster
and produces vectors better suited for tree-based estimators like
HistGradientBoostingClassifier. To do this, we can simply replace
the GapEncoder with the MinHashEncoder in the previous pipeline
using set_params().
from skrub import MinHashEncoder
minhash_pipe = make_pipeline(
TableVectorizer(high_cardinality=MinHashEncoder(n_components=30)),
HistGradientBoostingClassifier(),
)
minhash_results = cross_validate(minhash_pipe, X, y, scoring="roc_auc")
results.append(("MinHashEncoder", minhash_results))
plot_box_results(results)
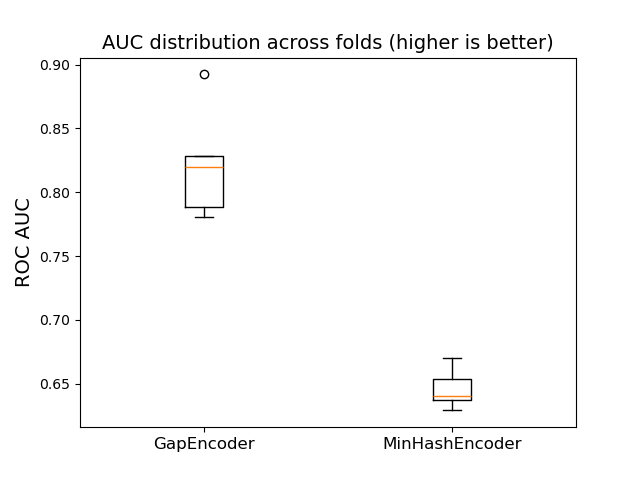
Remarkably, the vectors produced by the MinHashEncoder offer less predictive
power than those from the GapEncoder on this dataset.
TextEncoder#
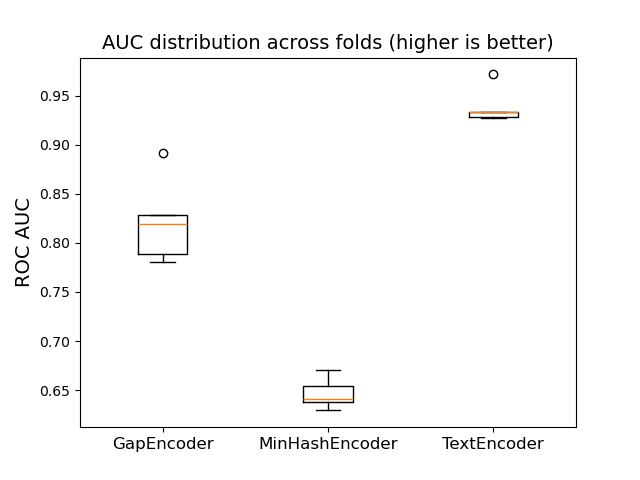
Let’s now shift our focus to pre-trained deep learning encoders. Our previous encoders are syntactic models that we trained directly on the toxicity dataset. To generate more powerful vector representations for free-form text and diverse entries, we can instead use semantic models, such as BERT, which have been trained on very large datasets.
TextEncoder enables you to integrate any Sentence Transformer model from the
Hugging Face Hub (or from your local disk) into your Pipeline to transform a text
column in a dataframe. By default, TextEncoder uses the e5-small-v2 model.
from skrub import TextEncoder
text_encoder = TextEncoder(
"sentence-transformers/paraphrase-albert-small-v2",
device="cpu",
)
text_encoder_pipe = make_pipeline(
TableVectorizer(high_cardinality=text_encoder),
HistGradientBoostingClassifier(),
)
text_encoder_results = cross_validate(text_encoder_pipe, X, y, scoring="roc_auc")
results.append(("TextEncoder", text_encoder_results))
plot_box_results(results)
StringEncoder#
TextEncoder embeddings are very strong, but they are also quite expensive to
use. A simpler, faster alternative for encoding strings is the StringEncoder,
which works by first performing a tf-idf (computing vectors of rescaled word
counts of the text wiki), and then
following it with TruncatedSVD to reduce the number of dimensions to, in this
case, 30.
from skrub import StringEncoder
string_encoder = StringEncoder(ngram_range=(3, 4), analyzer="char_wb", random_state=0)
string_encoder_pipe = make_pipeline(
TableVectorizer(high_cardinality=string_encoder),
HistGradientBoostingClassifier(),
)
string_encoder_results = cross_validate(string_encoder_pipe, X, y, scoring="roc_auc")
results.append(("StringEncoder", string_encoder_results))
plot_box_results(results)
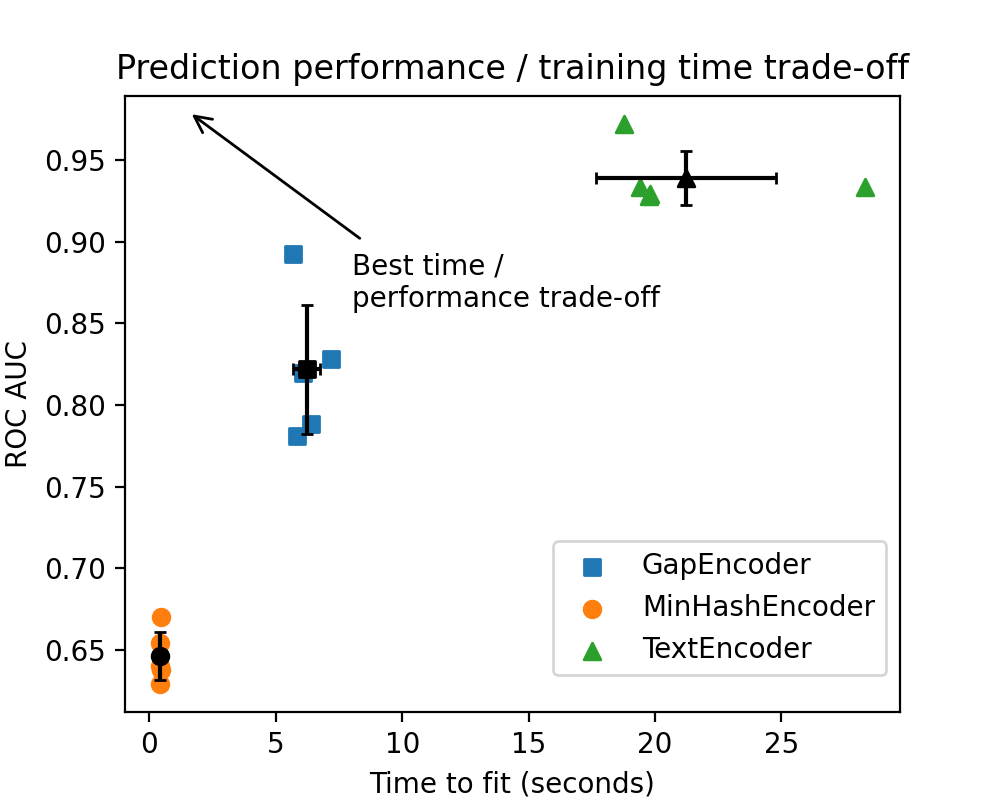
The performance of the TextEncoder is significantly stronger than that of
the syntactic encoders, which is expected. But how long does it take to load
and vectorize text on a CPU using a Sentence Transformer model? Below, we display
the tradeoff between predictive accuracy and training time. Note that since we are
not training the Sentence Transformer model, the “fitting time” refers to the
time taken for vectorization.
def plot_performance_tradeoff(results):
fig, ax = plt.subplots(figsize=(5, 4), dpi=200)
markers = ["s", "o", "^", "x"]
for idx, (name, result) in enumerate(results):
ax.scatter(
result["fit_time"],
result["test_score"],
label=name,
marker=markers[idx],
)
mean_fit_time = np.mean(result["fit_time"])
mean_score = np.mean(result["test_score"])
ax.scatter(
mean_fit_time,
mean_score,
color="k",
marker=markers[idx],
)
std_fit_time = np.std(result["fit_time"])
std_score = np.std(result["test_score"])
ax.errorbar(
x=mean_fit_time,
y=mean_score,
yerr=std_score,
fmt="none",
c="k",
capsize=2,
)
ax.errorbar(
x=mean_fit_time,
y=mean_score,
xerr=std_fit_time,
fmt="none",
c="k",
capsize=2,
)
ax.set_xscale("log")
ax.set_xlabel("Time to fit (seconds)")
ax.set_ylabel("ROC AUC")
ax.set_title("Prediction performance / training time trade-off")
ax.annotate(
"Best time / \nperformance trade-off",
xy=(0.05, 0.95),
xycoords="axes fraction",
xytext=(0.2, 0.8),
textcoords="axes fraction",
arrowprops=dict(arrowstyle="->", lw=1.5, mutation_scale=15),
)
ax.legend(bbox_to_anchor=(1.02, 0.3))
plt.show()
plot_performance_tradeoff(results)
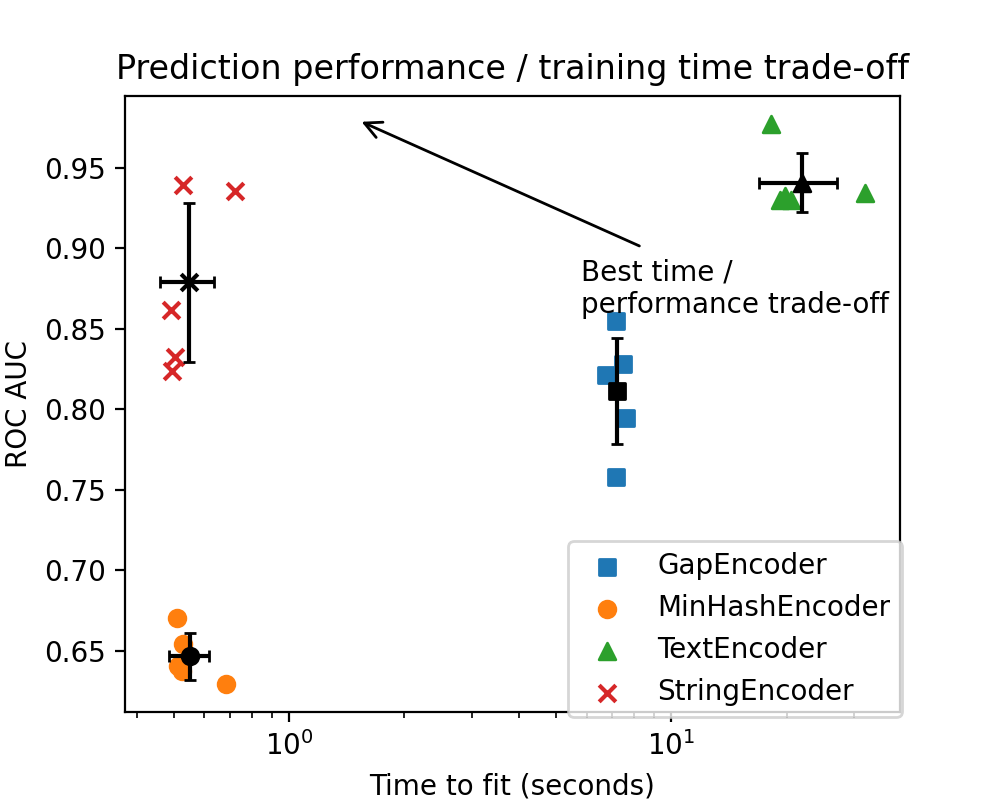
The black points represent the average time to fit and AUC for each vectorizer, and the width of the bars represents one standard deviation.
The green outlier dot on the right side of the plot corresponds to the first time the Sentence Transformers model was downloaded and loaded into memory. During the subsequent cross-validation iterations, the model is simply copied, which reduces computation time for the remaining folds.
Interestingly, StringEncoder has a performance remarkably similar to that of
GapEncoder, while being significantly faster.
Conclusion#
In conclusion, TextEncoder provides powerful vectorization for text, but at
the cost of longer computation times and the need for additional dependencies,
such as torch. StringEncoder represents a simpler alternative that can provide
good performance at a fraction of the cost of more complex methods.
Total running time of the script: (3 minutes 40.613 seconds)